Structural, Functional, and Immunological Characterization of Profilin Panallergens Amb a 8, Art v 4, and Bet v 2
- PMID: 27231348
- PMCID: PMC4957032
- DOI: 10.1074/jbc.M116.733659
Structural, Functional, and Immunological Characterization of Profilin Panallergens Amb a 8, Art v 4, and Bet v 2
Abstract
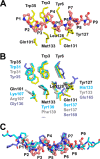
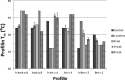
Ragweed allergens affect several million people in the United States and Canada. To date, only two ragweed allergens, Amb t 5 and Amb a 11, have their structures determined and deposited to the Protein Data Bank. Here, we present structures of methylated ragweed allergen Amb a 8, Amb a 8 in the presence of poly(l-proline), and Art v 4 (mugwort allergen). Amb a 8 and Art v 4 are panallergens belonging to the profilin family of proteins. They share significant sequence and structural similarities, which results in cross-recognition by IgE antibodies. Molecular and immunological properties of Amb a 8 and Art v 4 are compared with those of Bet v 2 (birch pollen allergen) as well as with other allergenic profilins. We purified recombinant allergens that are recognized by patient IgE and are highly cross-reactive. It was determined that the analyzed allergens are relatively unstable. Structures of Amb a 8 in complex with poly(l-proline)10 or poly(l-proline)14 are the first structures of the plant profilin in complex with proline-rich peptides. Amb a 8 binds the poly(l-proline) in a mode similar to that observed in human, mouse, and P. falciparum profilin·peptide complexes. However, only some of the residues that form the peptide binding site are conserved.
Keywords: IgE binding; allergen; allergy; crystal structure; epitope mapping; peptide binding; pollen; protein stability.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
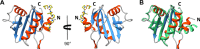

References
-
- Mari A. (2001) Multiple pollen sensitization: a molecular approach to the diagnosis. Int. Arch. Allergy Immunol. 125, 57–65 - PubMed
-
- Tinghino R., Twardosz A., Barletta B., Puggioni E. M., Iacovacci P., Butteroni C., Afferni C., Mari A., Hayek B., Di Felice G., Focke M., Westritschnig K., Valenta R., and Pini C. (2002) Molecular, structural, and immunologic relationships between different families of recombinant calcium-binding pollen allergens. J. Allergy Clin. Immunol. 109, 314–320 - PubMed
-
- Wopfner N., Gruber P., Wallner M., Briza P., Ebner C., Mari A., Richter K., Vogel L., and Ferreira F. (2008) Molecular and immunological characterization of novel weed pollen pan-allergens. Allergy 63, 872–881 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

