Epiregulin Recognition Mechanisms by Anti-epiregulin Antibody 9E5: STRUCTURAL, FUNCTIONAL, AND MOLECULAR DYNAMICS SIMULATION ANALYSES
- PMID: 26627827
- PMCID: PMC4732215
- DOI: 10.1074/jbc.M115.656009
Epiregulin Recognition Mechanisms by Anti-epiregulin Antibody 9E5: STRUCTURAL, FUNCTIONAL, AND MOLECULAR DYNAMICS SIMULATION ANALYSES
Abstract
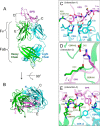
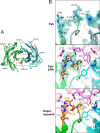
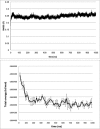
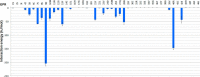
Epiregulin (EPR) is a ligand of the epidermal growth factor (EGF) family that upon binding to its epidermal growth factor receptor (EGFR) stimulates proliferative signaling, especially in colon cancer cells. Here, we describe the three-dimensional structure of the EPR antibody (the 9E5(Fab) fragment) in the presence and absence of EPR. Among the six complementarity-determining regions (CDRs), CDR1-3 in the light chain and CDR2 in the heavy chain predominantly recognize EPR. In particular, CDR3 in the heavy chain dramatically moves with cis-trans isomerization of Pro(103). A molecular dynamics simulation and mutational analyses revealed that Arg(40) in EPR is a key residue for the specific binding of 9E5 IgG. From isothermal titration calorimetry analysis, the dissociation constant was determined to be 6.5 nm. Surface plasmon resonance analysis revealed that the dissociation rate of 9E5 IgG is extremely slow. The superimposed structure of 9E5(Fab)·EPR on the known complex structure of EGF·EGFR showed that the 9E5(Fab) paratope overlaps with Domains I and III on the EGFR, which reveals that the 9E5(Fab)·EPR complex could not bind to the EGFR. The 9E5 antibody will also be useful in medicine as a neutralizing antibody specific for colon cancer.
Keywords: antibody; cancer; crystal structure; epidermal growth factor (EGF); molecular dynamics; thermodynamics.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




References
-
- Nielsen D. L., Kümler I., Palshof J. A., and Andersson M. (2013) Efficacy of HER2-targeted therapy in metastatic breast cancer. Monoclonal antibodies and tyrosine kinase inhibitors. Breast 22, 1–12 - PubMed
-
- Hynes N. E., and Lane H. A. (2005) ERBB receptors and cancer: the complexity of targeted inhibitors. Nat. Rev. Cancer 5, 341–354 - PubMed
-
- Yarden Y., and Sliwkowski M. X. (2001) Untangling the ErbB signalling network. Nat. Rev. Mol. Cell Biol. 2, 127–137 - PubMed
-
- Fischer O. M., Hart S., Gschwind A., and Ullrich A. (2003) EGFR signal transactivation in cancer cells. Biochem. Soc. Trans. 31, 1203–1208 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

