Structure and function of the BAH-containing domain of Orc1p in epigenetic silencing
- PMID: 12198162
- PMCID: PMC125411
- DOI: 10.1093/emboj/cdf468
Structure and function of the BAH-containing domain of Orc1p in epigenetic silencing
Abstract
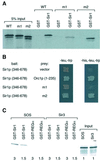
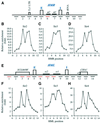
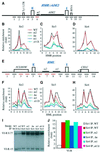
The N-terminal domain of the largest subunit of the Saccharomyces cerevisiae origin recognition complex (Orc1p) functions in transcriptional silencing and contains a bromo-adjacent homology (BAH) domain found in some chromatin-associated proteins including Sir3p. The 2.2 A crystal structure of the N-terminal domain of Orc1p revealed a BAH core and a non-conserved helical sub-domain. Mutational analyses demonstrated that the helical sub-domain was necessary and sufficient to bind Sir1p, and critical for targeting Sir1p primarily to the cis-acting E silencers at the HMR and HML silent chromatin domains. In the absence of the BAH domain, approximately 14-20% of cells in a population were silenced at the HML locus. Moreover, the distributions of the Sir2p, Sir3p and Sir4p proteins, while normal, were at levels lower than found in wild-type cells. Thus, in the absence of the Orc1p BAH domain, HML resembled silencing of genes adjacent to telomeres. These data are consistent with the view that the Orc1p-Sir1p interaction at the E silencers ensures stable inheritance of pre-established Sir2p, Sir3p and Sir4p complexes at the silent mating type loci.
Figures




References
-
- Bell S.P. and Stillman,B. (1992) ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature, 357, 128–134. - PubMed
-
- Bell S.P., Kobayashi,R. and Stillman,B. (1993) Yeast origin recognition complex functions in transcription silencing and DNA replication. Science, 262, 1844–1849. - PubMed
-
- Bell S.P., Mitchell,J., Leber,J., Kobayashi,R. and Stillman,B. (1995) The multidomain structure of Orc1p reveals similarity to regulators of DNA replication and transcriptional silencing. Cell, 83, 563–568. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

