ANKRD9 is a metabolically-controlled regulator of IMPDH2 abundance and macro-assembly
- PMID: 31337707
- PMCID: PMC6768640
- DOI: 10.1074/jbc.RA119.008231
ANKRD9 is a metabolically-controlled regulator of IMPDH2 abundance and macro-assembly
Abstract
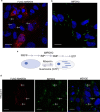
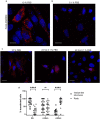
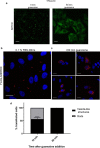
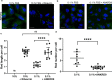
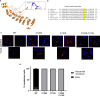
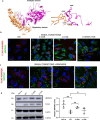
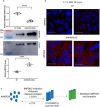
Members of a large family of Ankyrin Repeat Domain (ANKRD) proteins regulate numerous cellular processes by binding to specific protein targets and modulating their activity, stability, and other properties. The same ANKRD protein may interact with different targets and regulate distinct cellular pathways. The mechanisms responsible for switches in the ANKRDs' behavior are often unknown. We show that cells' metabolic state can markedly alter interactions of an ANKRD protein with its target and the functional outcomes of this interaction. ANKRD9 facilitates degradation of inosine monophosphate dehydrogenase 2 (IMPDH2), the rate-limiting enzyme in GTP biosynthesis. Under basal conditions ANKRD9 is largely segregated from the cytosolic IMPDH2 in vesicle-like structures. Upon nutrient limitation, ANKRD9 loses its vesicular pattern and assembles with IMPDH2 into rodlike filaments, in which IMPDH2 is stable. Inhibition of IMPDH2 activity with ribavirin favors ANKRD9 binding to IMPDH2 rods. The formation of ANKRD9/IMPDH2 rods is reversed by guanosine, which restores ANKRD9 associations with the vesicle-like structures. The conserved Cys109Cys110 motif in ANKRD9 is required for the vesicle-to-rods transition as well as binding and regulation of IMPDH2. Oppositely to overexpression, ANKRD9 knockdown increases IMPDH2 levels and prevents formation of IMPDH2 rods upon nutrient limitation. Taken together, the results suggest that a guanosine-dependent metabolic switch determines the mode of ANKRD9 action toward IMPDH2.
Keywords: ANKRD9; GTP; IMPDH2; ankyrin; ankyrin repeat domain; cell metabolism; complex; cytoophidia; nucleotide; protein-protein interaction; rod.
© 2019 Hayward et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures

References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

