Crystal structure of the M1 protein-binding domain of the influenza A virus nuclear export protein (NEP/NS2)
- PMID: 12970177
- PMCID: PMC212717
- DOI: 10.1093/emboj/cdg449
Crystal structure of the M1 protein-binding domain of the influenza A virus nuclear export protein (NEP/NS2)
Abstract
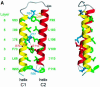
During influenza virus infection, viral ribonucleoproteins (vRNPs) are replicated in the nucleus and must be exported to the cytoplasm before assembling into mature viral particles. Nuclear export is mediated by the cellular protein Crm1 and putatively by the viral protein NEP/NS2. Proteolytic cleavage of NEP defines an N-terminal domain which mediates RanGTP-dependent binding to Crm1 and a C-terminal domain which binds to the viral matrix protein M1. The 2.6 A crystal structure of the C-terminal domain reveals an amphipathic helical hairpin which dimerizes as a four-helix bundle. The NEP-M1 interaction involves two critical epitopes: an exposed tryptophan (Trp78) surrounded by a cluster of glutamate residues on NEP, and the basic nuclear localization signal (NLS) of M1. Implications for vRNP export are discussed.
Figures









References
-
- Arzt S., Baudin,F., Barge,A., Timmins,P., Burmeister,W.P. and Ruigrok,R.W.H. (2001) Combined results from solution studies on intact influenza virus M1 protein and from a new crystal form of its N-terminal domain show that M1 is an elongated monomer. Virology, 279, 439–446. - PubMed
-
- Askjaer P., Heick Jensen,T., Nilsson,J., Engelmeier,L. and Kjems,J. (1998) The specificity of the CRM1-Rev nuclear export signal interaction is mediated by RanGTP. J. Biol. Chem., 273, 33414–33422. - PubMed
-
- Baudin F., Petit,I., Weissenhorn,W. and Ruigrok,R.W.H. (2001) In vitro dissection of the membrane binding and RNP binding activities of influenza virus M1 protein. Virology, 281, 102–108. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

