The C-terminal domain of DNA gyrase A adopts a DNA-bending beta-pinwheel fold
- PMID: 15123801
- PMCID: PMC409912
- DOI: 10.1073/pnas.0401595101
The C-terminal domain of DNA gyrase A adopts a DNA-bending beta-pinwheel fold
Abstract
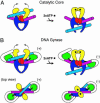
DNA gyrase is unique among enzymes for its ability to actively introduce negative supercoils into DNA. This function is mediated in part by the C-terminal domain of its A subunit (GyrA CTD). Here, we report the crystal structure of this approximately 35-kDa domain determined to 1.75-A resolution. The GyrA CTD unexpectedly adopts an unusual fold, which we term a beta-pinwheel, that is globally reminiscent of a beta-propeller but is built of blades with a previously unobserved topology. A large, conserved basic patch on the outer edge of this domain suggests a likely site for binding and bending DNA; fluorescence resonance energy transfer-based assays show that the GyrA CTD is capable of bending DNA by > or =180 degrees over a 40-bp region. Surprisingly, we find that the CTD of the topoisomerase IV A subunit, which shares limited sequence homology with the GyrA CTD, also bends DNA. Together, these data provide a physical explanation for the ability of DNA gyrase to constrain a positive superhelical DNA wrap, and also suggest that the particular substrate preferences of topoisomerase IV might be dictated in part by the function of this domain.
Figures
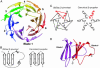

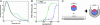
References
-
- Champoux, J. J. (2001) Annu. Rev. Biochem. 70, 369-413. - PubMed
-
- Wang, J. C. (2002) Nat. Rev. Mol. Cell Biol. 3, 430-440. - PubMed
-
- Hickson, I. D. (2003) Nat. Rev. Cancer 3, 169-178. - PubMed
-
- Brown, P. O. & Cozzarelli, N. R. (1979) Science 206, 1081-1083. - PubMed
-
- Liu, L. F., Liu, C. C. & Alberts, B. M. (1980) Cell 19, 697-707. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

