Structures of the Escherichia coli PutA proline dehydrogenase domain in complex with competitive inhibitors
- PMID: 15449943
- PMCID: PMC3727243
- DOI: 10.1021/bi048737e
Structures of the Escherichia coli PutA proline dehydrogenase domain in complex with competitive inhibitors
Abstract
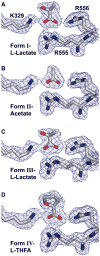
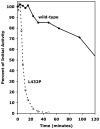
Proline dehydrogenase (PRODH) catalyzes the first step of proline catabolism, the flavin-dependent oxidation of proline to Delta(1)-pyrroline-5-carboxylate. Here we present a structure-based study of the PRODH active site of the multifunctional Escherichia coli proline utilization A (PutA) protein using X-ray crystallography, enzyme kinetic measurements, and site-directed mutagenesis. Structures of the PutA PRODH domain complexed with competitive inhibitors acetate (K(i) = 30 mM), L-lactate (K(i) = 1 mM), and L-tetrahydro-2-furoic acid (L-THFA, K(i) = 0.2 mM) have been determined to high-resolution limits of 2.1-2.0 A. The discovery of acetate as a competitive inhibitor suggests that the carboxyl is the minimum functional group recognized by the active site, and the structures show how the enzyme exploits hydrogen-bonding and nonpolar interactions to optimize affinity for the substrate. The PRODH/L-THFA complex is the first structure of PRODH with a five-membered ring proline analogue bound in the active site and thus provides new insights into substrate recognition and the catalytic mechanism. The ring of L-THFA is nearly parallel to the middle ring of the FAD isoalloxazine, with the inhibitor C5 atom 3.3 A from the FAD N5. This geometry suggests direct hydride transfer as a plausible mechanism. Mutation of conserved active site residue Leu432 to Pro caused a 5-fold decrease in k(cat) and a severe loss in thermostability. These changes are consistent with the location of Leu432 in the hydrophobic core near residues that directly contact FAD. Our results suggest that the molecular basis for increased plasma proline levels in schizophrenic subjects carrying the missense mutation L441P is due to decreased stability of human PRODH2.
Figures





References
-
- Phang JM. The Regulatory Functions of Proline and Pyrroline-5-Carboxylic Acid. Current Topics in Cell Regulation. 1985;25:92–132. - PubMed
-
- Adams E, Frank L. Metabolism of proline and the hydroxyprolines. Annu Rev Biochem. 1980;49:1005–61. - PubMed
-
- Donald SP, Sun XY, Hu CA, Yu J, Mei JM, Valle D, Phang JM. Proline oxidase, encoded by p53-induced gene-6, catalyzes the generation of proline-dependent reactive oxygen species. Cancer Res. 2001;61:1810–5. - PubMed
-
- Polyak K, Xia Y, Zweier JL, Kinzler KW, Vogelstein B. A model for p53-induced apoptosis. Nature. 1997;389:300–5. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

