Structure of the receptor-binding domain of human thrombopoietin determined by complexation with a neutralizing antibody fragment
- PMID: 14769915
- PMCID: PMC357010
- DOI: 10.1073/pnas.0308530100
Structure of the receptor-binding domain of human thrombopoietin determined by complexation with a neutralizing antibody fragment
Abstract
The cytokine thrombopoietin (TPO), the ligand for the hematopoietic receptor c-Mpl, acts as a primary regulator of megakaryocytopoiesis and platelet production. We have determined the crystal structure of the receptor-binding domain of human TPO (hTPO(163)) to a 2.5-A resolution by complexation with a neutralizing Fab fragment. The backbone structure of hTPO(163) has an antiparallel four-helix bundle fold. The neutralizing Fab mainly recognizes the C-D crossover loop containing the species invariant residue Q111. Titration calorimetric experiments show that hTPO(163) interacts with soluble c-Mpl containing the extracellular cytokine receptor homology domains with 1:2 stoichiometry with the binding constants of 3.3 x 10(9) M(-1) and 1.1 x 10(6) M(-1). The presence of the neutralizing Fab did not inhibit binding of hTPO(163) to soluble c-Mpl fragments, but the lower-affinity binding disappeared. Together with prior genetic data, these define the structure-function relationships in TPO and the activation scheme of c-Mpl.
Figures

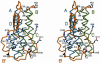


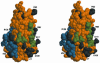
References
-
- Kelemen, E., Cserhati, I. & Tanos, B. (1958) Acta Haematol. 20, 350–355. - PubMed
-
- Bartley, T. D., Bogenberger, J., Hunt, P., Li, Y. S., Lu, H. S., Martin, F., Chang, M. S., Samal, B., Nichol, J. L., Swift, S., et al. (1994) Cell 77, 1117–1124. - PubMed
-
- de Sauvage, F. J., Hass, P. E., Spencer, S. D., Malloy, B. E., Gurney, A. L., Spencer, S. A., Darbonne, W. C., Henzel, W. J., Wong, S. C., Kuang, W.-J., et al. (1994) Nature 369, 533–538. - PubMed
-
- Lok, S., Kaushansky, K., Holly, R. D., Kuijper, J. L., Lofton-Day, C. E., Oort, P. J., Grant, F. J., Heipel, M. D., Burkhead, S. K., Kramer, J. M., et al. (1994) Nature 369, 565–568. - PubMed
-
- Kato, T., Ogami, K., Shimada, Y., Iwamatsu, A., Sohma, Y., Akahori, H., Horie, K., Kokubo, A., Kudo, Y., Maeda, E., et al. (1995) J. Biochem. 118, 229–236, and correction (1995) 119, 208. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

