A divergent external loop confers antagonistic activity on floral regulators FT and TFL1
- PMID: 16424903
- PMCID: PMC1383534
- DOI: 10.1038/sj.emboj.7600950
A divergent external loop confers antagonistic activity on floral regulators FT and TFL1
Abstract
The Arabidopsis genes FT and TERMINAL FLOWER1 (TFL1) encode related proteins with similarity to human Raf kinase inhibitor protein. FT, and likely also TFL1, is recruited to the promoters of floral genes through interaction with FD, a bZIP transcription factor. FT, however, induces flowering, while TFL1 represses flowering. Residues responsible for the opposite activities of FT and TFL1 were mapped by examining plants that overexpress chimeric proteins. A region important in vivo localizes to a 14-amino-acid segment that evolves very rapidly in TFL1 orthologs, but is almost invariant in FT orthologs. Crystal structures show that this segment forms an external loop of variable conformation. The only residue unambiguously distinguishing the FT and TFL1 loops makes a hydrogen bond with a residue near the entrance of a potential ligand-binding pocket in TFL1, but not in FT. This pocket is contacted by a C-terminal peptide, which also contributes to the opposite FT and TFL1 activities. In combination, these results identify a molecular surface likely to be recognized by FT- and/or TFL1-specific interactors.
Figures



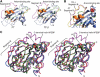
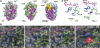


References
-
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056 - PubMed
-
- Alvarez J, Guli CL, Yu X-H, Smyth DR (1992) Terminal flower: a gene affecting inflorescence development in Arabidopsis thaliana. Plant J 2: 103–116
-
- Ausubel FM, Brent R, Kingston RZ, Moore DD, Seideman JG, Smith JA, Struhl K (1991) Current Protocols in Molecular Biology. New York, NY: Wiley
-
- Banfield MJ, Barker JJ, Perry AC, Brady RL (1998) Function from structure? The crystal structure of human phosphatidylethanolamine-binding protein suggests a role in membrane signal transduction. Structure 6: 1245–1254 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

