Structural analysis and solution studies of the activated regulatory domain of the response regulator ArcA: a symmetric dimer mediated by the alpha4-beta5-alpha5 face
- PMID: 15876365
- PMCID: PMC3690759
- DOI: 10.1016/j.jmb.2005.03.059
Structural analysis and solution studies of the activated regulatory domain of the response regulator ArcA: a symmetric dimer mediated by the alpha4-beta5-alpha5 face
Abstract
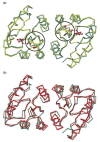
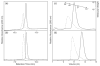
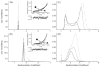
Escherichia coli react to changes from aerobic to anaerobic conditions of growth using the ArcA-ArcB two-component signal transduction system. This system, in conjunction with other proteins, regulates the respiratory metabolic pathways in the organism. ArcA is a member of the OmpR/PhoB subfamily of response regulator transcription factors that are known to regulate transcription by binding in tandem to target DNA direct repeats. It is still unclear in this subfamily how activation by phosphorylation of the regulatory domain of response regulators stimulates DNA binding by the effector domain and how dimerization and domain orientation, as well as intra- and intermolecular interactions, affect this process. In order to address these questions we have solved the crystal structures of the regulatory domain of ArcA in the presence and absence of the phosphoryl analog, BeF3-. In the crystal structures, the regulatory domain of ArcA forms a symmetric dimer mediated by the alpha4-beta5-alpha5 face of the protein and involving a number of residues that are highly conserved in the OmpR/PhoB subfamily. It is hypothesized that members of this subfamily use a common mechanism of regulation by dimerization. Additional biophysical studies were employed to probe the oligomerization state of ArcA, as well as its individual domains, in solution. The solution studies show the propensity of the individual domains to associate into oligomers larger than the dimer observed for the intact protein, and suggest that the C-terminal DNA-binding domain also plays a role in oligomerization.
Figures


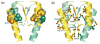


References
-
- Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. - PubMed
-
- West AH, Stock AM. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem Sci. 2001;26:369–376. - PubMed
-
- Macielag MJ, Goldschmidt R. Inhibitors of bacterial two-component signalling systems. Expert Opin Investig Drugs. 2000;9:2351–2369. - PubMed
-
- Stephenson K, Hoch JA. Developing inhibitors to selectively target two-component and phosphorelay signal transduction systems of pathogenic microorganisms. Curr Med Chem. 2004;11:765–773. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

