Impact of amino acid substitutions at secondary structures in the BRCT domains of the tumor suppressor BRCA1: Implications for clinical annotation
- PMID: 30765603
- PMCID: PMC6463708
- DOI: 10.1074/jbc.RA118.005274
Impact of amino acid substitutions at secondary structures in the BRCT domains of the tumor suppressor BRCA1: Implications for clinical annotation
Abstract
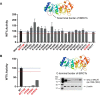
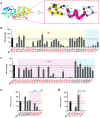
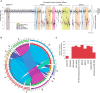
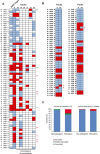
Genetic testing for BRCA1, a DNA repair protein, can identify carriers of pathogenic variants associated with a substantially increased risk for breast and ovarian cancers. However, an association with increased risk is unclear for a large fraction of BRCA1 variants present in the human population. Most of these variants of uncertain clinical significance lead to amino acid changes in the BRCA1 protein. Functional assays are valuable tools to assess the potential pathogenicity of these variants. Here, we systematically probed the effects of substitutions in the C terminus of BRCA1: the N- and C-terminal borders of its tandem BRCT domain, the BRCT-[N-C] linker region, and the α1 and α'1 helices in BRCT-[N] and -[C]. Using a validated transcriptional assay based on a fusion of the GAL4 DNA-binding domain to the BRCA1 C terminus (amino acids 1396-1863), we assessed the functional impact of 99 missense variants of BRCA1. We include the data obtained for these 99 missense variants in a joint analysis to generate the likelihood of pathogenicity for 347 missense variants in BRCA1 using VarCall, a Bayesian integrative statistical model. The results from this analysis increase our understanding of BRCA1 regions less tolerant to changes, identify functional borders of structural domains, and predict the likelihood of pathogenicity for 98% of all BRCA1 missense variants in this region recorded in the population. This knowledge will be critical for improving risk assessment and clinical treatment of carriers of BRCA1 variants.
Keywords: BRCA1; BRCT domains; VarCall; breast cancer; breast cancer risk; cancer prevention; clinical annotation; clinical risk assessment; human genetics; protein conformation.
© 2019 Fernandes et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures


References
-
- Mavaddat N., Peock S., Frost D., Ellis S., Platte R., Fineberg E., Evans D. G., Izatt L., Eeles R. A., Adlard J., Davidson R., Eccles D., Cole T., et al. (2013) Cancer risks for BRCA1 and BRCA2 mutation carriers: results from prospective analysis of EMBRACE. J. Natl. Cancer Inst. 105, 812–822 10.1093/jnci/djt095 - DOI - PubMed
-
- Frank T. S., Deffenbaugh A. M., Reid J. E., Hulick M., Ward B. E., Lingenfelter B., Gumpper K. L., Scholl T., Tavtigian S. V., Pruss D. R., and Critchfield G. C. (2002) Clinical characteristics of individuals with germline mutations in BRCA1 and BRCA2: analysis of 10,000 individuals. J. Clin. Oncol. 20, 1480–1490 10.1200/JCO.2002.20.6.1480 - DOI - PubMed
-
- Goldgar D. E., Easton D. F., Deffenbaugh A. M., Monteiro A. N., Tavtigian S. V., and Couch F. J., Breast Cancer Information Core (BIC) Steering Committee (2004) Integrated evaluation of DNA sequence variants of unknown clinical significance: application to BRCA1 and BRCA2. Am. J. Hum. Genet. 75, 535–544 10.1086/424388 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

