Crystal structure of prostate-specific membrane antigen, a tumor marker and peptidase
- PMID: 15837926
- PMCID: PMC556220
- DOI: 10.1073/pnas.0502101102
Crystal structure of prostate-specific membrane antigen, a tumor marker and peptidase
Abstract
Prostate-specific membrane antigen (PSMA) is highly expressed in prostate cancer cells and nonprostatic solid tumor neovasculature and is a target for anticancer imaging and therapeutic agents. PSMA acts as a glutamate carboxypeptidase (GCPII) on small molecule substrates, including folate, the anticancer drug methotrexate, and the neuropeptide N-acetyl-l-aspartyl-l-glutamate. Here we present the 3.5-A crystal structure of the PSMA ectodomain, which reveals a homodimer with structural similarity to transferrin receptor, a receptor for iron-loaded transferrin that lacks protease activity. Unlike transferrin receptor, the protease domain of PSMA contains a binuclear zinc site, catalytic residues, and a proposed substrate-binding arginine patch. Elucidation of the PSMA structure combined with docking studies and a proposed catalytic mechanism provides insight into the recognition of inhibitors and the natural substrate N-acetyl-l-aspartyl-l-glutamate. The PSMA structure will facilitate development of chemotherapeutics, cancer-imaging agents, and agents for treatment of neurological disorders.
Figures

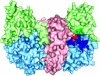

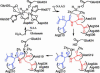
References
-
- Ghosh, A. & Heston, W. D. W. (2004) J. Cell. Biochem. 91, 528-539. - PubMed
-
- Pangalos, M. N., Neefs, J. M., Somers, M., Verhasselt, P., Bekkers, M., van der Helm, L., Fraiponts, E., Ashton, D. & Gordon, R. D. (1999) J. Biol. Chem. 274, 8470-8483. - PubMed
-
- Berger, U. V., Luthi-Carter, R., Passani, L. A., Elkabes, S., Black, I., Konradi, C. & Coyle, J. T. (1999) J. Comp. Neurol. 415, 52-64. - PubMed
-
- Gong, M. C., Chang, S. S., Sadelain, M., Bander, N. H. & Heston, W. D. W. (1999) Cancer Metastasis Rev. 18, 483-490. - PubMed
-
- Elgamal, A. A., Holmes, E. H., Su, S. L., Tino, W. T., Simmons, S. J., Peterson, M., Greene, T. G., Boynton, A. L. & Murphy, G. P. (2000) Semin. Surg. Oncol. 18, 10-16. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

