Kinetic and Structural Impact of Metal Ions and Genetic Variations on Human DNA Polymerase ι
- PMID: 27555320
- PMCID: PMC5076516
- DOI: 10.1074/jbc.M116.748285
Kinetic and Structural Impact of Metal Ions and Genetic Variations on Human DNA Polymerase ι
Abstract
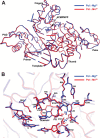
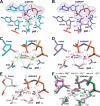
DNA polymerase (pol) ι is a Y-family polymerase involved in translesion synthesis, exhibiting higher catalytic activity with Mn2+ than Mg2+ The human germline R96G variant impairs both Mn2+-dependent and Mg2+-dependent activities of pol ι, whereas the Δ1-25 variant selectively enhances its Mg2+-dependent activity. We analyzed pre-steady-state kinetic and structural effects of these two metal ions and genetic variations on pol ι using pol ι core (residues 1-445) proteins. The presence of Mn2+ (0.15 mm) instead of Mg2+ (2 mm) caused a 770-fold increase in efficiency (kpol/Kd,dCTP) of pol ι for dCTP insertion opposite G, mainly due to a 450-fold decrease in Kd,dCTP The R96G and Δ1-25 variants displayed a 53-fold decrease and a 3-fold increase, respectively, in kpol/Kd,dCTP for dCTP insertion opposite G with Mg2+ when compared with wild type, substantially attenuated by substitution with Mn2+ Crystal structures of pol ι ternary complexes, including the primer terminus 3'-OH and a non-hydrolyzable dCTP analogue opposite G with the active-site Mg2+ or Mn2+, revealed that Mn2+ achieves more optimal octahedral coordination geometry than Mg2+, with lower values in average coordination distance geometry in the catalytic metal A-site. Crystal structures of R96G revealed the loss of three H-bonds of residues Gly-96 and Tyr-93 with an incoming dNTP, due to the lack of an arginine, as well as a destabilized Tyr-93 side chain secondary to the loss of a cation-π interaction between both side chains. These results provide a mechanistic basis for alteration in pol ι catalytic function with coordinating metals and genetic variation.
Keywords: DNA enzyme; DNA polymerase; X-ray crystallography; crystal structure; enzyme kinetics; genetic variation; kinetics; metal ion-protein interaction; pre-steady-state kinetics; translesion DNA synthesis.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

References
-
- Loeb L. A., and Monnat R. J. Jr. (2008) DNA polymerases and human disease. Nat. Rev. Genet. 9, 594–604 - PubMed
-
- Ling H., Boudsocq F., Woodgate R., and Yang W. (2001) Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell 107, 91–102 - PubMed
-
- Choi J.-Y., Eoff R. E., and Guengerich F. P. (2011) Bypass DNA polymerases. in Chemical Carcinogenesis (Penning T. M., ed), pp. 345–373, Humana Press, New York, NY
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

