Recognition of a functional peroxisome type 1 target by the dynamic import receptor pex5p
- PMID: 17157249
- PMCID: PMC5030714
- DOI: 10.1016/j.molcel.2006.10.024
Recognition of a functional peroxisome type 1 target by the dynamic import receptor pex5p
Abstract
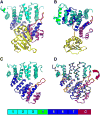
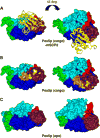
Peroxisomes require the translocation of folded and functional target proteins of various sizes across the peroxisomal membrane. We have investigated the structure and function of the principal import receptor Pex5p, which recognizes targets bearing a C-terminal peroxisomal targeting signal type 1. Crystal structures of the receptor in the presence and absence of a peroxisomal target, sterol carrier protein 2, reveal major structural changes from an open, snail-like conformation into a closed, circular conformation. These changes are caused by a long loop C terminal to the 7-fold tetratricopeptide repeat segments. Mutations in residues of this loop lead to defects in peroxisomal import in human fibroblasts. The structure of the receptor/cargo complex demonstrates that the primary receptor-binding site of the cargo is structurally and topologically autonomous, enabling the cargo to retain its native structure and function.
Figures



References
-
- Agne B, Meindl NM, Niederhoff K, Einwachter H, Rehling P, Sickmann A, Meyer HE, Girzalsky W, Kunau WH. Pex8p: an intraperoxisomal organizer of the peroxisomal import machinery. Mol Cell. 2003;11:635–646. - PubMed
-
- Battiste JL, Wagner G. Utilization of site-directed spin labeling and high-resolution heteronuclear nuclear magnetic resonance for global fold determination of large proteins with limited nuclear overhauser effect data. Biochemistry. 2000;39:5355–5365. - PubMed
-
- Braverman N, Dodt G, Gould SJ, Valle D. An isoform of Pex5p, the human PTS1 receptor, is required for the import of PTS2 proteins into peroxisomes. Hum Mol Genet. 1998;7:1195–1205. - PubMed
-
- Brocard CB, Jedeszko C, Song HC, Terlecky SR, Walton PA. Protein structure and import into the peroxisomal matrix. Traffic. 2003;4:74–82. - PubMed
-
- Brown NP, Leroy C, Sander C. MView: a web-compatible database search or multiple alignment viewer. Bioinformatics. 1998;14:380–381. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

