Structure of a putative lipoate protein ligase from Thermoplasma acidophilum and the mechanism of target selection for post-translational modification
- PMID: 16384580
- PMCID: PMC7610907
- DOI: 10.1016/j.jmb.2005.11.057
Structure of a putative lipoate protein ligase from Thermoplasma acidophilum and the mechanism of target selection for post-translational modification
Abstract
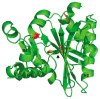
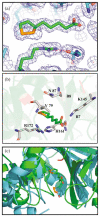
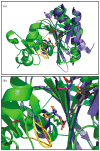
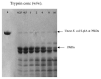
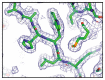
Lipoyl-lysine swinging arms are crucial to the reactions catalysed by the 2-oxo acid dehydrogenase multienzyme complexes. A gene encoding a putative lipoate protein ligase (LplA) of Thermoplasma acidophilum was cloned and expressed in Escherichia coli. The recombinant protein, a monomer of molecular mass 29 kDa, was catalytically inactive. Crystal structures in the absence and presence of bound lipoic acid were solved at 2.1 A resolution. The protein was found to fall into the alpha/beta class and to be structurally homologous to the catalytic domains of class II aminoacyl-tRNA synthases and biotin protein ligase, BirA. Lipoic acid in LplA was bound in the same position as biotin in BirA. The structure of the T.acidophilum LplA and limited proteolysis of E.coli LplA together highlighted some key features of the post-translational modification. A loop comprising residues 71-79 in the T.acidophilum ligase is proposed as interacting with the dithiolane ring of lipoic acid and discriminating against the entry of biotin. A second loop comprising residues 179-193 was disordered in the T.acidophilum structure; tryptic cleavage of the corresponding loop in the E.coli LplA under non-denaturing conditions rendered the enzyme catalytically inactive, emphasizing its importance. The putative LplA of T.acidophilum lacks a C-terminal domain found in its counterparts in E.coli (Gram-negative) or Streptococcus pneumoniae (Gram-positive). A gene encoding a protein that appears to have structural homology to the additional domain in the E.coli and S.pneumoniae enzymes was detected alongside the structural gene encoding the putative LplA in the T.acidophilum genome. It is likely that this protein is required to confer activity on the LplA as currently purified, one protein perhaps catalysing the formation of the obligatory lipoyl-AMP intermediate, and the other transferring the lipoyl group from it to the specific lysine residue in the target protein.
Figures



References
-
- Perham RN. Swinging arms and swinging domains in multifunctional enzymes: catalytic machines for multistep reactions. Annu Rev Biochem. 2000;69:961–1004. - PubMed
-
- de Kok A, Hengeveld AF, Martin A, Westphal AH. The pyruvate dehydrogenase multienzyme complex from Gram-negative bacteria. Bio-chim BioPhys Acta. 1998;1385:353–366. - PubMed
-
- Motokawa Y, Fujiwara K, Okamura-Ikeda K. in Health and Disease. In: Packer L, Cadenas E, editors. Biothiols in Health and Disease. Marcel Dekker; New York: 1995. pp. 389–407.
-
- Reed LJ, Hackert ML. Structure-function relationships in dihydrolipoamide acyltransferases. J Biol Chem. 1990;265:8971–8974. - PubMed
-
- Perham RN. Domains, motifs and linkers in 2-oxo acid dehydrogenase multienzyme complexes: a paradigm in the design of a multifunctional enzyme. Biochemistry. 1991;30:8501–8512. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

