Polyadenylation factor CPSF-73 is the pre-mRNA 3'-end-processing endonuclease
- PMID: 17128255
- PMCID: PMC3866582
- DOI: 10.1038/nature05363
Polyadenylation factor CPSF-73 is the pre-mRNA 3'-end-processing endonuclease
Abstract
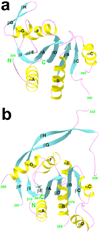
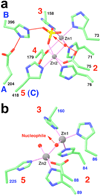
Most eukaryotic messenger RNA precursors (pre-mRNAs) undergo extensive maturational processing, including cleavage and polyadenylation at the 3'-end. Despite the characterization of many proteins that are required for the cleavage reaction, the identity of the endonuclease is not known. Recent analyses indicated that the 73-kDa subunit of cleavage and polyadenylation specificity factor (CPSF-73) might be the endonuclease for this and related reactions, although no direct data confirmed this. Here we report the crystal structures of human CPSF-73 at 2.1 A resolution, complexed with zinc ions and a sulphate that might mimic the phosphate group of the substrate, and the related yeast protein CPSF-100 (Ydh1) at 2.5 A resolution. Both CPSF-73 and CPSF-100 contain two domains, a metallo-beta-lactamase domain and a novel beta-CASP (named for metallo-beta-lactamase, CPSF, Artemis, Snm1, Pso2) domain. The active site of CPSF-73, with two zinc ions, is located at the interface of the two domains. Purified recombinant CPSF-73 possesses RNA endonuclease activity, and mutations that disrupt zinc binding in the active site abolish this activity. Our studies provide the first direct experimental evidence that CPSF-73 is the pre-mRNA 3'-end-processing endonuclease.
Figures


References
-
- Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. - PubMed
-
- Wahle E, Ruegsegger U. 3'-end processing of pre-mRNA in eukaryotes. FEMS Microbiol. Rev. 1999;23:277–295. - PubMed
-
- Proudfoot NJ. New perspectives on connecting messenger RNA 3' end formation to transcription. Curr. Opin. Cell Biol. 2004;16:272–278. - PubMed
-
- Zorio DAR, Bentley D. The link between mRNA processing and transcription: communication works both ways. Exp. Cell Res. 2004;296:91–97. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

