A community resource of experimental data for NMR / X-ray crystal structure pairs
- PMID: 26293815
- PMCID: PMC4815321
- DOI: 10.1002/pro.2774
A community resource of experimental data for NMR / X-ray crystal structure pairs
Abstract
We have developed an online NMR / X-ray Structure Pair Data Repository. The NIGMS Protein Structure Initiative (PSI) has provided many valuable reagents, 3D structures, and technologies for structural biology. The Northeast Structural Genomics Consortium was one of several PSI centers. NESG used both X-ray crystallography and NMR spectroscopy for protein structure determination. A key goal of the PSI was to provide experimental structures for at least one representative of each of hundreds of targeted protein domain families. In some cases, structures for identical (or nearly identical) constructs were determined by both NMR and X-ray crystallography. NMR spectroscopy and X-ray diffraction data for 41 of these "NMR / X-ray" structure pairs determined using conventional triple-resonance NMR methods with extensive sidechain resonance assignments have been organized in an online NMR / X-ray Structure Pair Data Repository. In addition, several NMR data sets for perdeuterated, methyl-protonated protein samples are included in this repository. As an example of the utility of this repository, these data were used to revisit questions about the precision and accuracy of protein NMR structures first outlined by Levy and coworkers several years ago (Andrec et al., Proteins 2007;69:449-465). These results demonstrate that the agreement between NMR and X-ray crystal structures is improved using modern methods of protein NMR spectroscopy. The NMR / X-ray Structure Pair Data Repository will provide a valuable resource for new computational NMR methods development.
Keywords: X-ray crystallography; accuracy and precision of NMR structures; protein NMR spectroscopy; structural bioinformatics.
© 2015 The Protein Society.
Figures




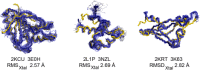

References
-
- Montelione GT, Anderson S (1999) Structural genomics: keystone for a Human Proteome Project. Nat Struct Biol 6:11–12. - PubMed
-
- Liu J, Montelione GT, Rost B (2007) Novel leverage of structural genomics. Nat Biotechnol 25:849–851. - PubMed
-
- Graslund S, Nordlund P, Weigelt J, Hallberg BM, Bray J, Gileadi O, Knapp S, Oppermann U, Arrowsmith C, Hui R, Ming J, dhe‐Paganon S, Park HW, Savchenko A, Yee A, Edwards A, Vincentelli R, Cambillau C, Kim R, Kim SH, Rao Z, Shi Y, Terwilliger TC, Kim CY, Hung LW, Waldo GS, Peleg Y, Albeck S, Unger T, Dym O, Prilusky J, Sussman JL, Stevens RC, Lesley SA, Wilson IA, Joachimiak A, Collart F, Dementieva I, Donnelly MI, Eschenfeldt WH, Kim Y, Stols L, Wu R, Zhou M, Burley SK, Emtage JS, Sauder JM, Thompson D, Bain K, Luz J, Gheyi T, Zhang F, Atwell S, Almo SC, Bonanno JB, Fiser A, Swaminathan S, Studier FW, Chance MR, Sali A, Acton TB, Xiao R, Zhao L, Ma LC, Hunt JF, Tong L, Cunningham K, Inouye M, Anderson S, Janjua H, Shastry R, Ho CK, Wang D, Wang H, Jiang M, Montelione GT, Stuart DI, Owens RJ, Daenke S, Schutz A, Heinemann U, Yokoyama S, Bussow K, Gunsalus KC (2008) Protein production and purification. Nat Methods 5:135–146. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

