Structural biology of the IL-1 superfamily: key cytokines in the regulation of immune and inflammatory responses
- PMID: 24677376
- PMCID: PMC4005705
- DOI: 10.1002/pro.2441
Structural biology of the IL-1 superfamily: key cytokines in the regulation of immune and inflammatory responses
Abstract
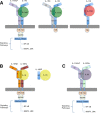
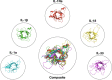
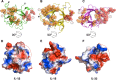
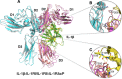
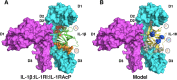
Interleukin-1 superfamily of cytokines (IL-1, IL-18, IL-33) play key roles in inflammation and regulating immunity. The mechanisms of agonism and antagonism in the IL-1 superfamily have been pursued by structural biologists for nearly 20 years. New insights into these mechanisms were recently provided by the crystal structures of the ternary complexes of IL-1β and its receptors. We will review here the structural biology related to receptor recognition by IL-1 superfamily cytokines and the regulation of its cytokine activities by antagonists.
Keywords: agonist; antagonist; cytokine; inflammation; interleukin; receptors.
© 2014 The Protein Society.
Figures


References
-
- Dinarello C, Arend W, Sims J, Smith D, Blumberg H, O'Neill L, Goldbach-Mansky R, Pizarro T, Hoffman H, Bufler P, Nold M, Ghezzi P, Mantovani A, Garlanda C, Boraschi D, Rubartelli A, Netea M, van der Meer J, Joosten L, Mandrup-Poulsen T, Donath M, Lewis E, Pfeilschifter J, Martin M, Kracht M, Muehl H, Novick D, Lukic M, Conti B, Solinger A, Kelk P, van de Veerdonk F, Gabel C. IL-1 family nomenclature. Nat Immunol. 2010;11:973. - PMC - PubMed
-
- Dinarello CA. Immunological and inflammatory functions of the interleukin-1 family. Annu Rev Immunol. 2009;27:519–550. - PubMed
-
- Arend WP, Palmer G, Gabay C. IL-1, IL-18, and IL-33 families of cytokines. Immunol Rev. 2008;223:20–38. - PubMed
-
- O'Neill LA. The interleukin-1 receptor/Toll-like receptor superfamily: 10 years of progress. Immunol Rev. 2008;226:10–18. - PubMed
-
- Heguy A, Baldari CT, Macchia G, Telford JL, Melli M. Amino acids conserved in interleukin-1 receptors (IL-1Rs) and the Drosophila toll protein are essential for IL-1R signal transduction. J Biol Chem. 1992;267:2605–2609. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

