The chemokine X-factor: Structure-function analysis of the CXC motif at CXCR4 and ACKR3
- PMID: 32788219
- PMCID: PMC7535910
- DOI: 10.1074/jbc.RA120.014244
The chemokine X-factor: Structure-function analysis of the CXC motif at CXCR4 and ACKR3
Abstract
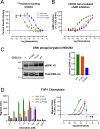
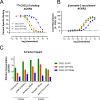
The human chemokine family consists of 46 protein ligands that induce chemotactic cell migration by activating a family of 23 G protein-coupled receptors. The two major chemokine subfamilies, CC and CXC, bind distinct receptor subsets. A sequence motif defining these families, the X position in the CXC motif, is not predicted to make significant contacts with the receptor, but instead links structural elements associated with binding and activation. Here, we use comparative analysis of chemokine NMR structures, structural modeling, and molecular dynamic simulations that suggested the X position reorients the chemokine N terminus. Using CXCL12 as a model CXC chemokine, deletion of the X residue (Pro-10) had little to no impact on the folded chemokine structure but diminished CXCR4 agonist activity as measured by ERK phosphorylation, chemotaxis, and Gi/o-mediated cAMP inhibition. Functional impairment was attributed to over 100-fold loss of CXCR4 binding affinity. Binding to the other CXCL12 receptor, ACKR3, was diminished nearly 500-fold. Deletion of Pro-10 had little effect on CXCL12 binding to the CXCR4 N terminus, a major component of the chemokine-GPCR interface. Replacement of the X residue with the most frequent amino acid at this position (P10Q) had an intermediate effect between WT and P10del in each assay, with ACKR3 having a higher tolerance for this mutation. This work shows that the X residue helps to position the CXCL12 N terminus for optimal docking into the orthosteric pocket of CXCR4 and suggests that the CC/CXC motif contributes directly to receptor selectivity by orienting the chemokine N terminus in a subfamily-specific direction.
Keywords: CC/CXC motif; G protein-coupled receptor (GPCR); cell migration; chemokine; chemokine network; nuclear magnetic resonance (NMR); signal transduction; structure-function.
© 2020 Wedemeyer et al.
Conflict of interest statement
Conflict of interest—B. F. V. and F. C. P. have ownership interests in Protein Foundry, LLC.
Figures




References
-
- Bachelerie F., Ben-Baruch A., Burkhardt A. M., Combadiere C., Farber J. M., Graham G. J., Horuk R., Sparre-Ulrich A. H., Locati M., Luster A. D., Mantovani A., Matsushima K., Murphy P. M., Nibbs R., Nomiyama H., et al. (2014) International Union of Pharmacology. LXXXIX. Update on the extended family of chemokine receptors and introducing a new nomenclature for atypical chemokine receptors. Pharmacol. Rev. 66, 1–79 10.1124/pr.113.007724 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

