Structure of the Brd4 ET domain bound to a C-terminal motif from γ-retroviral integrases reveals a conserved mechanism of interaction
- PMID: 26858406
- PMCID: PMC4776507
- DOI: 10.1073/pnas.1516813113
Structure of the Brd4 ET domain bound to a C-terminal motif from γ-retroviral integrases reveals a conserved mechanism of interaction
Abstract
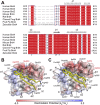
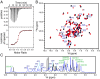
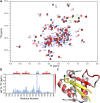
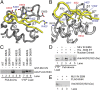
The bromodomain and extraterminal domain (BET) protein family are promising therapeutic targets for a range of diseases linked to transcriptional activation, cancer, viral latency, and viral integration. Tandem bromodomains selectively tether BET proteins to chromatin by engaging cognate acetylated histone marks, and the extraterminal (ET) domain is the focal point for recruiting a range of cellular and viral proteins. BET proteins guide γ-retroviral integration to transcription start sites and enhancers through bimodal interaction with chromatin and the γ-retroviral integrase (IN). We report the NMR-derived solution structure of the Brd4 ET domain bound to a conserved peptide sequence from the C terminus of murine leukemia virus (MLV) IN. The complex reveals a protein-protein interaction governed by the binding-coupled folding of disordered regions in both interacting partners to form a well-structured intermolecular three-stranded β sheet. In addition, we show that a peptide comprising the ET binding motif (EBM) of MLV IN can disrupt the cognate interaction of Brd4 with NSD3, and that substitutions of Brd4 ET residues essential for binding MLV IN also impair interaction of Brd4 with a number of cellular partners involved in transcriptional regulation and chromatin remodeling. This suggests that γ-retroviruses have evolved the EBM to mimic a cognate interaction motif to achieve effective integration in host chromatin. Collectively, our findings identify key structural features of the ET domain of Brd4 that allow for interactions with both cellular and viral proteins.
Keywords: BET family transcription factor; ET binding motif (EBM); NMR; protein–protein interaction; retroviral integration.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- McBride AA, McPhillips MG, Oliveira JG. Brd4: Tethering, segregation and beyond. Trends Microbiol. 2004;12(12):527–529. - PubMed
-
- Wu SY, Chiang CM. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J Biol Chem. 2007;282(18):13141–13145. - PubMed
-
- Yang Z, et al. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19(4):535–545. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

