Structural Basis for the Discriminative Recognition of N6-Methyladenosine RNA by the Human YT521-B Homology Domain Family of Proteins
- PMID: 26318451
- PMCID: PMC4598999
- DOI: 10.1074/jbc.M115.680389
Structural Basis for the Discriminative Recognition of N6-Methyladenosine RNA by the Human YT521-B Homology Domain Family of Proteins
Abstract
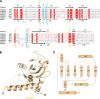
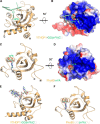
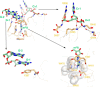
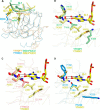
N(6)-Methyladenosine (m(6)A) is the most abundant internal modification in RNA and is specifically recognized by YT521-B homology (YTH) domain-containing proteins. Recently we reported that YTHDC1 prefers guanosine and disfavors adenosine at the position preceding the m(6)A nucleotide in RNA and preferentially binds to the GG(m(6)A)C sequence. Now we systematically characterized the binding affinities of the YTH domains of three other human proteins and yeast YTH domain protein Pho92 and determined the crystal structures of the YTH domains of human YTHDF1 and yeast Pho92 in complex with a 5-mer m(6)A RNA, respectively. Our binding and structural data revealed that the YTH domain used a conserved aromatic cage to recognize m(6)A. Nevertheless, none of these YTH domains, except YTHDC1, display sequence selectivity at the position preceding the m(6)A modification. Structural comparison of these different YTH domains revealed that among those, only YTHDC1 harbors a distinctly selective binding pocket for the nucleotide preceding the m(6)A nucleotide.
Keywords: N6-methyladenosine; RNA binding protein; RNA methylation; RNA-protein interaction; crystal structure; isothermal titration calorimetry (ITC).
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


References
-
- Kouzarides T. (2007) Chromatin modifications and their function. Cell 128, 693–705 - PubMed
-
- Machnicka M. A., Milanowska K., Osman Oglou O., Purta E., Kurkowska M., Olchowik A., Januszewski W., Kalinowski S., Dunin-Horkawicz S., Rother K. M., Helm M., Bujnicki J. M., Grosjean H. (2013) MODOMICS: a database of RNA modification pathways: 2013 update. Nucleic Acids Res. 41, D262–D267 - PMC - PubMed
-
- Bokar J. A. (2005) Fine-tuning of RNA Functions by Modification and Editing (Grosjean H., ed) pp. 141–177, Springer-Verlag, Berlin
-
- Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M., Sorek R., Rechavi G. (2012) Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

