Modulation of androgen receptor activation function 2 by testosterone and dihydrotestosterone
- PMID: 17591767
- PMCID: PMC4075031
- DOI: 10.1074/jbc.M703268200
Modulation of androgen receptor activation function 2 by testosterone and dihydrotestosterone
Abstract
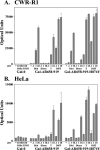
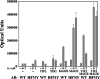
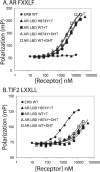
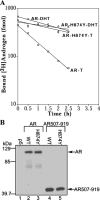
The androgen receptor (AR) is transcriptionally activated by high affinity binding of testosterone (T) or its 5alpha-reduced metabolite, dihydrotestosterone (DHT), a more potent androgen required for male reproductive tract development. The molecular basis for the weaker activity of T was investigated by determining T-bound ligand binding domain crystal structures of wild-type AR and a prostate cancer somatic mutant complexed with the AR FXXLF or coactivator LXXLL peptide. Nearly identical interactions of T and DHT in the AR ligand binding pocket correlate with similar rates of dissociation from an AR fragment containing the ligand binding domain. However, T induces weaker AR FXXLF and coactivator LXXLL motif interactions at activation function 2 (AF2). Less effective FXXLF motif binding to AF2 accounts for faster T dissociation from full-length AR. T can nevertheless acquire DHT-like activity through an AR helix-10 H874Y prostate cancer mutation. The Tyr-874 mutant side chain mediates a new hydrogen bonding scheme from exterior helix-10 to backbone protein core helix-4 residue Tyr-739 to rescue T-induced AR activity by improving AF2 binding of FXXLF and LXXLL motifs. Greater AR AF2 activity by improved core helix interactions is supported by the effects of melanoma antigen gene protein-11, an AR coregulator that binds the AR FXXLF motif and targets AF2 for activation. We conclude that T is a weaker androgen than DHT because of less favorable T-dependent AR FXXLF and coactivator LXXLL motif interactions at AF2.
Figures






References
-
- Wilson EM, French FS. J Biol Chem. 1976;251:5620–5629. - PubMed
-
- Simental JA, Sar M, Lane MV, French FS, Wilson EM. J Biol Chem. 1991;266:510–518. - PubMed
-
- Warnmark A, Treuter E, Wright AP, Gustafsson JA. Mol Endocrinol. 2003;17:1901–1909. - PubMed
-
- He B, Gampe RT, Kole AJ, Hnat AT, Stanley TB, An G, Stewart EL, Kalman RI, Minges JT, Wilson EM. Mol Cell. 2004;16:425–438. - PubMed
-
- He B, Kemppainen JA, Wilson EM. J Biol Chem. 2000;275:22986–22994. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

