A Combination of Structural and Empirical Analyses Delineates the Key Contacts Mediating Stability and Affinity Increases in an Optimized Biotherapeutic Single-chain Fv (scFv)
- PMID: 26515064
- PMCID: PMC4714214
- DOI: 10.1074/jbc.M115.688010
A Combination of Structural and Empirical Analyses Delineates the Key Contacts Mediating Stability and Affinity Increases in an Optimized Biotherapeutic Single-chain Fv (scFv)
Abstract
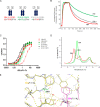
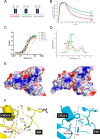
Fully-human single-chain Fv (scFv) proteins are key potential building blocks of bispecific therapeutic antibodies, but they often suffer from manufacturability and clinical development limitations such as instability and aggregation. The causes of these scFv instability problems, in proteins that should be theoretically stable, remains poorly understood. To inform the future development of such molecules, we carried out a comprehensive structural analysis of the highly stabilized anti-CXCL13 scFv E10. E10 was derived from the parental 3B4 using complementarity-determining region (CDR)-restricted mutagenesis and tailored selection and screening strategies, and carries four mutations in VL-CDR3. High-resolution crystal structures of parental 3B4 and optimized E10 scFvs were solved in the presence and absence of human CXCL13. In parallel, a series of scFv mutants was generated to interrogate the individual contribution of each of the four mutations to stability and affinity improvements. In combination, these analyses demonstrated that the optimization of E10 was primarily mediated by removing clashes between both the VL and the VH, and between the VL and CXCL13. Importantly, a single, germline-encoded VL-CDR3 residue mediated the key difference between the stable and unstable forms of the scFv. This work demonstrates that, aside from being the critical mediators of specificity and affinity, CDRs may also be the primary drivers of biotherapeutic developability.
Keywords: antibody engineering; chemokine; crystal structure; mutagenesis in vitro; protein stability.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



References
-
- Chan A. C., and Carter P. J. (2010) Therapeutic antibodies for autoimmunity and inflammation. Nat. Rev. Immunol. 10, 301–316 - PubMed
-
- Bargou R., Leo E., Zugmaier G., Klinger M., Goebeler M., Knop S., Noppeney R., Viardot A., Hess G., Schuler M., Einsele H., Brandl C., Wolf A., Kirchinger P., Klappers P., Schmidt M., Riethmüller G., Reinhardt C., Baeuerle P. A., and Kufer P. (2008) Tumor regression in cancer patients by very low doses of a T cell-engaging antibody. Science 321, 974–977 - PubMed
-
- Fitzgerald J. B., Johnson B. W., Baum J., Adams S., Iadevaia S., Tang J., Rimkunas V., Xu L., Kohli N., Rennard R., Razlog M., Jiao Y., Harms B. D., Olivier K. J. Jr., Schoeberl B., Nielsen U. B., and Lugovskoy A. A. (2014) MM-141, an IGF-IR- and ErbB3-directed bispecific antibody, overcomes network adaptations that limit activity of IGF-IR inhibitors. Mol. Cancer Ther. 13, 410–425 - PubMed
-
- Liddy N., Bossi G., Adams K. J., Lissina A., Mahon T. M., Hassan N. J., Gavarret J., Bianchi F. C., Pumphrey N. J., Ladell K., Gostick E., Sewell A. K., Lissin N. M., Harwood N. E., Molloy P. E., Li Y., Cameron B. J., Sami M., Baston E. E., Todorov P. T., Paston S. J., Dennis R. E., Harper J. V., Dunn S. M., Ashfield R., Johnson A., McGrath Y., Plesa G., June C. H., Kalos M., Price D. A., Vuidepot A., Williams D. D., Sutton D. H., and Jakobsen B. K. (2012) Monoclonal TCR-redirected tumor cell killing. Nat. Med. 18, 980–987 - PubMed
-
- Moore P. A., Zhang W., Rainey G. J., Burke S., Li H., Huang L., Gorlatov S., Veri M. C., Aggarwal S., Yang Y., Shah K., Jin L., Zhang S., He L., Zhang T., Ciccarone V., Koenig S., Bonvini E., and Johnson S. (2011) Application of dual affinity retargeting molecules to achieve optimal redirected T-cell killing of B-cell lymphoma. Blood 117, 4542–4551 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

