Activation segment dimerization: a mechanism for kinase autophosphorylation of non-consensus sites
- PMID: 18239682
- PMCID: PMC2239268
- DOI: 10.1038/emboj.2008.8
Activation segment dimerization: a mechanism for kinase autophosphorylation of non-consensus sites
Abstract
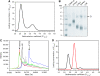
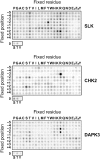
Protein kinase autophosphorylation of activation segment residues is a common regulatory mechanism in phosphorylation-dependent signalling cascades. However, the molecular mechanisms that guarantee specific and efficient phosphorylation of these sites have not been elucidated. Here, we report on three novel and diverse protein kinase structures that reveal an exchanged activation segment conformation. This dimeric arrangement results in an active kinase conformation in trans, with activation segment phosphorylation sites in close proximity to the active site of the interacting protomer. Analytical ultracentrifugation and chemical cross-linking confirmed the presence of dimers in solution. Consensus substrate sequences for each kinase showed that the identified activation segment autophosphorylation sites are non-consensus substrate sites. Based on the presented structural and functional data, a model for specific activation segment phosphorylation at non-consensus substrate sites is proposed that is likely to be common to other kinases from diverse subfamilies.
Figures




References
-
- Ahn JY, Li X, Davis HL, Canman CE (2002) Phosphorylation of threonine 68 promotes oligomerization and autophosphorylation of the Chk2 protein kinase via the forkhead-associated domain. J Biol Chem 277: 19389–19395 - PubMed
-
- Bignell G, Smith R, Hunter C, Stephens P, Davies H, Greenman C, Teague J, Butler A, Edkins S, Stevens C, O'Meara S, Parker A, Avis T, Barthorpe S, Brackenbury L, Buck G, Clements J, Cole J, Dicks E, Edwards K et al. (2006) Sequence analysis of the protein kinase gene family in human testicular germ-cell tumors of adolescents and adults. Genes Chromosomes Cancer 45: 42–46 - PMC - PubMed
-
- Brown NR, Noble ME, Lawrie AM, Morris MC, Tunnah P, Divita G, Johnson LN, Endicott JA (1999) Effects of phosphorylation of threonine 160 on cyclin-dependent kinase 2 structure and activity. J Biol Chem 274: 8746–8756 - PubMed
-
- Burakov AV, Kovalenko OV, Potekhina ES, Nadezhdina ES, Zinovkina LA (2005) LOSK (SLK) protein kinase activity is necessary for microtubule organization in the interphase cell centrosome. Dokl Biol Sci 403: 317–319 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

