Crystal structures of barley thioredoxin h isoforms HvTrxh1 and HvTrxh2 reveal features involved in protein recognition and possibly in discriminating the isoform specificity
- PMID: 18424513
- PMCID: PMC2386739
- DOI: 10.1110/ps.083460308
Crystal structures of barley thioredoxin h isoforms HvTrxh1 and HvTrxh2 reveal features involved in protein recognition and possibly in discriminating the isoform specificity
Abstract
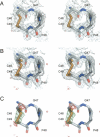
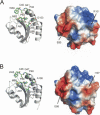
H-type thioredoxins (Trxs) constitute a particularly large Trx sub-group in higher plants. Here, the crystal structures are determined for the two barley Trx h isoforms, HvTrxh1 and HvTrxh2, in the partially radiation-reduced state to resolutions of 1.7 A, and for HvTrxh2 in the oxidized state to 2.0 A. The two Trxs have a sequence identity of 51% and highly similar fold and active-site architecture. Interestingly, the four independent molecules in the crystals of HvTrxh1 form two relatively large and essentially identical protein-protein interfaces. In each interface, a loop segment of one HvTrxh1 molecule is positioned along a shallow hydrophobic groove at the primary nucleophile Cys40 of another HvTrxh1 molecule. The association mode can serve as a model for the target protein recognition by Trx, as it brings the Met82 Cgamma atom (gamma position as a disulfide sulfur) of the bound loop segment in the proximity of the Cys40 thiol. The interaction involves three characteristic backbone-backbone hydrogen bonds in an antiparallel beta-sheet-like arrangement, similar to the arrangement observed in the structure of an engineered, covalently bound complex between Trx and a substrate protein, as reported by Maeda et al. in an earlier paper. The occurrence of an intermolecular salt bridge between Glu80 of the bound loop segment and Arg101 near the hydrophobic groove suggests that charge complementarity plays a role in the specificity of Trx. In HvTrxh2, isoleucine corresponds to this arginine, which emphasizes the potential for specificity differences between the coexisting barley Trx isoforms.
Figures



References
-
- Arnér, E.S.J., Holmgren, A. Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. Biochem. 2000;267:6102–6109. - PubMed
-
- Bahadur, R.P., Chakrabarti, P., Rodier, F., Janin, J. Dissecting subunit interfaces in homodimeric proteins. Proteins. 2003;53:708–719. - PubMed
-
- Brünger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. - PubMed
-
- Buchanan, B.B., Balmer, Y. Redox regulation: A broadening horizon. Annu. Rev. Plant Biol. 2005;56:187–220. - PubMed
-
- Capitani, G., Markovic-Housley, Z., DelVal, G., Morris, M., Jansonius, J.N., Schurmann, P. Crystal structures of two functionally different thioredoxins in spinach chloroplasts. J. Mol. Biol. 2000;302:135–154. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

