Molecular Basis of Filtering Carbapenems by Porins from β-Lactam-resistant Clinical Strains of Escherichia coli
- PMID: 26645688
- PMCID: PMC4742748
- DOI: 10.1074/jbc.M115.690156
Molecular Basis of Filtering Carbapenems by Porins from β-Lactam-resistant Clinical Strains of Escherichia coli
Abstract
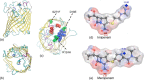
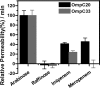
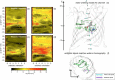
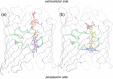
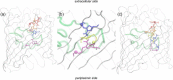
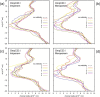
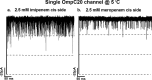
Integral membrane proteins known as porins are the major pathway by which hydrophilic antibiotics cross the outer membrane of Gram-negative bacteria. Single point mutations in porins can decrease the permeability of an antibiotic, either by reduction of channel size or modification of electrostatics in the channel, and thereby confer clinical resistance. Here, we investigate four mutant OmpC proteins from four different clinical isolates of Escherichia coli obtained sequentially from a single patient during a course of antimicrobial chemotherapy. OmpC porin from the first isolate (OmpC20) undergoes three consecutive and additive substitutions giving rise to OmpC26, OmpC28, and finally OmpC33. The permeability of two zwitterionic carbapenems, imipenem and meropenem, measured using liposome permeation assays and single channel electrophysiology differs significantly between OmpC20 and OmpC33. Molecular dynamic simulations show that the antibiotics must pass through the constriction zone of porins with a specific orientation, where the antibiotic dipole is aligned along the electric field inside the porin. We identify that changes in the vector of the electric field in the mutated porin, OmpC33, create an additional barrier by "trapping" the antibiotic in an unfavorable orientation in the constriction zone that suffers steric hindrance for the reorientation needed for its onward translocation. Identification and understanding the underlying molecular details of such a barrier to translocation will aid in the design of new antibiotics with improved permeation properties in Gram-negative bacteria.
Keywords: Porin; antibiotic resistance; electrophysiology; gram-negative bacteria; membrane biophysics; membrane transport; metadynamics; molecular dynamics simulations.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

References
-
- Wright G. D. (2007) The antibiotic resistome: the nexus of chemical and genetic diversity. Nat. Rev. Microbiol. 5, 175–186 - PubMed
-
- Levy S. B., and Marshall B. (2004) Antibacterial resistance worldwide: causes, challenges and responses. Nat. Med. 10, S122–9 - PubMed
-
- Payne D. J., Gwynn M. N., Holmes D. J., and Pompliano D. L. (2007) Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 6, 29–40 - PubMed
-
- Stavenger R. A., and Winterhalter M. (2014) TRANSLOCATION project: how to get good drugs into bad bugs. Sci. Transl. Med. 6, 228ed7 - PubMed
-
- Nakae T. (1976) Outer membrane of Salmonella: isolation of protein complex that produces transmembrane channels. J. Biol. Chem. 251, 2176–2178 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources

