Major Variations in HIV-1 Capsid Assembly Morphologies Involve Minor Variations in Molecular Structures of Structurally Ordered Protein Segments
- PMID: 27129282
- PMCID: PMC4933226
- DOI: 10.1074/jbc.M116.720557
Major Variations in HIV-1 Capsid Assembly Morphologies Involve Minor Variations in Molecular Structures of Structurally Ordered Protein Segments
Abstract
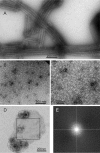
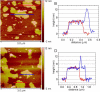
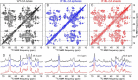
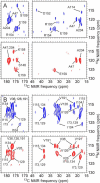
We present the results of solid state nuclear magnetic resonance (NMR) experiments on HIV-1 capsid protein (CA) assemblies with three different morphologies, namely wild-type CA (WT-CA) tubes with 35-60 nm diameters, planar sheets formed by the Arg(18)-Leu mutant (R18L-CA), and R18L-CA spheres with 20-100 nm diameters. The experiments are intended to elucidate molecular structural variations that underlie these variations in CA assembly morphology. We find that multidimensional solid state NMR spectra of (15)N,(13)C-labeled CA assemblies are remarkably similar for the three morphologies, with only small differences in (15)N and (13)C chemical shifts, no significant differences in NMR line widths, and few differences in the number of detectable NMR cross-peaks. Thus, the pronounced differences in morphology do not involve major differences in the conformations and identities of structurally ordered protein segments. Instead, morphological variations are attributable to variations in conformational distributions within disordered segments, which do not contribute to the solid state NMR spectra. Variations in solid state NMR signals from certain amino acid side chains are also observed, suggesting differences in the intermolecular dimerization interface between curved and planar CA lattices, as well as possible differences in intramolecular helix-helix packing.
Keywords: human immunodeficiency virus (HIV); molecular dynamics; protein assembly; protein structure; solid state NMR.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures





Comment on
-
Structure of full-length HIV-1 CA: a model for the mature capsid lattice.Cell. 2007 Oct 5;131(1):70-9. doi: 10.1016/j.cell.2007.08.018. Cell. 2007. PMID: 17923088
References
-
- Ganser-Pornillos B. K., Cheng A., and Yeager M. (2007) Structure of full-length HIV-1 CA: a model for the mature capsid lattice. Cell 131, 70–79 - PubMed
-
- Li S., Hill C. P., Sundquist W. I., and Finch J. T. (2000) Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature 407, 409–413 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

