Structural basis for the function and inhibition of an influenza virus proton channel
- PMID: 18235504
- PMCID: PMC3889492
- DOI: 10.1038/nature06528
Structural basis for the function and inhibition of an influenza virus proton channel
Erratum in
- Nature. 2008 Mar 20;452(7185):380
Abstract
The M2 protein from influenza A virus is a pH-activated proton channel that mediates acidification of the interior of viral particles entrapped in endosomes. M2 is the target of the anti-influenza drugs amantadine and rimantadine; recently, resistance to these drugs in humans, birds and pigs has reached more than 90% (ref. 1). Here we describe the crystal structure of the transmembrane-spanning region of the homotetrameric protein in the presence and absence of the channel-blocking drug amantadine. pH-dependent structural changes occur near a set of conserved His and Trp residues that are involved in proton gating. The drug-binding site is lined by residues that are mutated in amantadine-resistant viruses. Binding of amantadine physically occludes the pore, and might also perturb the pK(a) of the critical His residue. The structure provides a starting point for solving the problem of resistance to M2-channel blockers.
Conflict of interest statement
Conflict of interest: WFD was the scientific founder of InfluMedix.
Figures
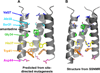
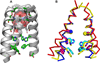

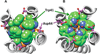
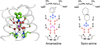
Comment in
-
Ion channels: coughing up flu's proton channels.Nature. 2008 Jan 31;451(7178):532-3. doi: 10.1038/451532a. Nature. 2008. PMID: 18235492 No abstract available.
References
-
- Gandhi CS, Shuck K, Lear JD, Dieckmann GR, DeGrado WF, Lamb RA, Pinto LH. Cu(II) inhibition of the proton translocation machinery of the influenza A virus M2 protein. J Biol Chem. 1999;274:5474–5482. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

