Crystal structure of full-length KcsA in its closed conformation
- PMID: 19346472
- PMCID: PMC2672561
- DOI: 10.1073/pnas.0810663106
Crystal structure of full-length KcsA in its closed conformation
Abstract
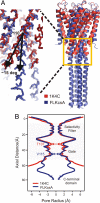
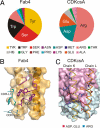
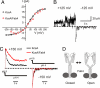
KcsA is a proton-activated, voltage-modulated K(+) channel that has served as the archetype pore domain in the Kv channel superfamily. Here, we have used synthetic antigen-binding fragments (Fabs) as crystallographic chaperones to determine the structure of full-length KcsA at 3.8 A, as well as that of its isolated C-terminal domain at 2.6 A. The structure of the full-length KcsA-Fab complex reveals a well-defined, 4-helix bundle that projects approximately 70 A toward the cytoplasm. This bundle promotes a approximately 15 degree bending in the inner bundle gate, tightening its diameter and shifting the narrowest point 2 turns of helix below. Functional analysis of the full-length KcsA-Fab complex suggests that the C-terminal bundle remains whole during gating. We suggest that this structure likely represents the physiologically relevant closed conformation of KcsA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Hille B. Ion Channels of Excitable Membranes. 3rd Ed. Sunderland, MA: Sinauer; 2003.
-
- Doyle DA, et al. The structure of the potassium channel: Molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. - PubMed
-
- Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution. Nature. 2001;414:43–48. - PubMed
-
- Kelly BL, Gross A. Potassium channel gating observed with site-directed mass tagging. Nat Struct Biol. 2003;10:280–284. - PubMed
-
- Perozo E, Cortes DM, Cuello LG. Structural rearrangements underlying K+-channel activation gating. Science. 1999;285:73–78. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

