Adaptive protein evolution grants organismal fitness by improving catalysis and flexibility
- PMID: 19098096
- PMCID: PMC2634896
- DOI: 10.1073/pnas.0807989106
Adaptive protein evolution grants organismal fitness by improving catalysis and flexibility
Abstract
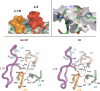
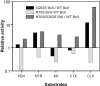
Protein evolution is crucial for organismal adaptation and fitness. This process takes place by shaping a given 3-dimensional fold for its particular biochemical function within the metabolic requirements and constraints of the environment. The complex interplay between sequence, structure, functionality, and stability that gives rise to a particular phenotype has limited the identification of traits acquired through evolution. This is further complicated by the fact that mutations are pleiotropic, and interactions between mutations are not always understood. Antibiotic resistance mediated by beta-lactamases represents an evolutionary paradigm in which organismal fitness depends on the catalytic efficiency of a single enzyme. Based on this, we have dissected the structural and mechanistic features acquired by an optimized metallo-beta-lactamase (MbetaL) obtained by directed evolution. We show that antibiotic resistance mediated by this enzyme is driven by 2 mutations with sign epistasis. One mutation stabilizes a catalytically relevant intermediate by fine tuning the position of 1 metal ion; whereas the other acts by augmenting the protein flexibility. We found that enzyme evolution (and the associated antibiotic resistance) occurred at the expense of the protein stability, revealing that MbetaLs have not exhausted their stability threshold. Our results demonstrate that flexibility is an essential trait that can be acquired during evolution on stable protein scaffolds. Directed evolution aided by a thorough characterization of the selected proteins can be successfully used to predict future evolutionary events and design inhibitors with an evolutionary perspective.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

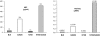
References
-
- Poelwijk FJ, Kiviet DJ, Weinreich DM, Tans SJ. Empirical fitness landscapes reveal accessible evolutionary paths. Nature. 2007;445:383–386. - PubMed
-
- Depristo MA, Weinreich DM, Hartl DL. Missense meanderings in sequence space: A biophysical view of protein evolution. Nat Rev Genet. 2005;6:678–687. - PubMed
-
- Bershtein S, Segal M, Bekerman R, Tokuriki N, Tawfik DS. Robustness-epistasis link shapes the fitness landscape of a randomly drifting protein. Nature. 2006;444:929–932. - PubMed
-
- Weinreich DM, Delaney NF, Depristo MA, Hartl DL. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science. 2006;312:111–114. - PubMed
-
- Orencia MC, Yoon JS, Ness JE, Stemmer WP, Stevens RC. Predicting the emergence of antibiotic resistance by directed evolution and structural analysis. Nat Struct Biol. 2001;8:238–242. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

