X-ray structures of the hexameric building block of the HIV capsid
- PMID: 19523676
- PMCID: PMC2840706
- DOI: 10.1016/j.cell.2009.04.063
X-ray structures of the hexameric building block of the HIV capsid
Abstract
The mature capsids of HIV and other retroviruses organize and package the viral genome and its associated enzymes for delivery into host cells. The HIV capsid is a fullerene cone: a variably curved, closed shell composed of approximately 250 hexamers and exactly 12 pentamers of the viral CA protein. We devised methods for isolating soluble, assembly-competent CA hexamers and derived four crystallographically independent models that define the structure of this capsid assembly unit at atomic resolution. A ring of six CA N-terminal domains form an apparently rigid core, surrounded by an outer ring of C-terminal domains. Mobility of the outer ring appears to be an underlying mechanism for generating the variably curved lattice in authentic capsids. Hexamer-stabilizing interfaces are highly hydrated, and this property may be key to the formation of quasi-equivalent interactions within hexamers and pentamers. The structures also clarify the molecular basis for capsid assembly inhibition and should facilitate structure-based drug design strategies.
Figures


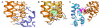

References
-
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. - PubMed
-
- Bartonova V, Igonet S, Sticht J, Glass B, Habermann A, Vaney MC, Sehr P, Lewis J, Rey FA, Kräusslich HG. Residues in the HIV-1 capsid assembly inhibitor binding site are essential for maintaining the assembly-competent quaternary structure of the capsid protein. J Biol Chem. 2008;283:32024–32033. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

