RNF8 E3 Ubiquitin Ligase Stimulates Ubc13 E2 Conjugating Activity That Is Essential for DNA Double Strand Break Signaling and BRCA1 Tumor Suppressor Recruitment
- PMID: 26903517
- PMCID: PMC4850281
- DOI: 10.1074/jbc.M116.715698
RNF8 E3 Ubiquitin Ligase Stimulates Ubc13 E2 Conjugating Activity That Is Essential for DNA Double Strand Break Signaling and BRCA1 Tumor Suppressor Recruitment
Abstract
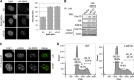
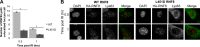
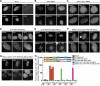
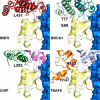
DNA double strand break (DSB) responses depend on the sequential actions of the E3 ubiquitin ligases RNF8 and RNF168 plus E2 ubiquitin-conjugating enzyme Ubc13 to specifically generate histone Lys-63-linked ubiquitin chains in DSB signaling. Here, we defined the activated RNF8-Ubc13∼ubiquitin complex by x-ray crystallography and its functional solution conformations by x-ray scattering, as tested by separation-of-function mutations imaged in cells by immunofluorescence. The collective results show that the RING E3 RNF8 targets E2 Ubc13 to DSB sites and plays a critical role in damage signaling by stimulating polyubiquitination through modulating conformations of ubiquitin covalently linked to the Ubc13 active site. Structure-guided separation-of-function mutations show that the RNF8 E2 stimulating activity is essential for DSB signaling in mammalian cells and is necessary for downstream recruitment of 53BP1 and BRCA1. Chromatin-targeted RNF168 rescues 53BP1 recruitment involved in non-homologous end joining but not BRCA1 recruitment for homologous recombination. These findings suggest an allosteric approach to targeting the ubiquitin-docking cleft at the E2-E3 interface for possible interventions in cancer and chronic inflammation, and moreover, they establish an independent RNF8 role in BRCA1 recruitment.
Keywords: 53BP1; BRCA1; DNA damage response; E3 ubiquitin-protein ligase RNF8 (RNF8); RNF168; Ubc13; cell biology; ubiquitylation (ubiquitination); x-ray crystallography; x-ray scattering.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures






References
-
- Pickart C. M., and Eddins M. J. (2004) Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta 1695, 55–72 - PubMed
-
- Haas A. L., and Siepmann T. J. (1997) Pathways of ubiquitin conjugation. FASEB J. 11, 1257–1268 - PubMed
-
- Dye B. T., and Schulman B. A. (2007) Structural mechanisms underlying posttranslational modification by ubiquitin-like proteins. Annu. Rev. Biophys. Biomol. Struct. 36, 131–150 - PubMed
-
- Komander D., and Rape M. (2012) The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

