4.0-A resolution cryo-EM structure of the mammalian chaperonin TRiC/CCT reveals its unique subunit arrangement
- PMID: 20194787
- PMCID: PMC2841888
- DOI: 10.1073/pnas.0913774107
4.0-A resolution cryo-EM structure of the mammalian chaperonin TRiC/CCT reveals its unique subunit arrangement
Abstract
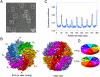
The essential double-ring eukaryotic chaperonin TRiC/CCT (TCP1-ring complex or chaperonin containing TCP1) assists the folding of approximately 5-10% of the cellular proteome. Many TRiC substrates cannot be folded by other chaperonins from prokaryotes or archaea. These unique folding properties are likely linked to TRiC's unique heterooligomeric subunit organization, whereby each ring consists of eight different paralogous subunits in an arrangement that remains uncertain. Using single particle cryo-EM without imposing symmetry, we determined the mammalian TRiC structure at 4.7-A resolution. This revealed the existence of a 2-fold axis between its two rings resulting in two homotypic subunit interactions across the rings. A subsequent 2-fold symmetrized map yielded a 4.0-A resolution structure that evinces the densities of a large fraction of side chains, loops, and insertions. These features permitted unambiguous identification of all eight individual subunits, despite their sequence similarity. Independent biochemical near-neighbor analysis supports our cryo-EM derived TRiC subunit arrangement. We obtained a Calpha backbone model for each subunit from an initial homology model refined against the cryo-EM density. A subsequently optimized atomic model for a subunit showed approximately 95% of the main chain dihedral angles in the allowable regions of the Ramachandran plot. The determination of the TRiC subunit arrangement opens the way to understand its unique function and mechanism. In particular, an unevenly distributed positively charged wall lining the closed folding chamber of TRiC differs strikingly from that of prokaryotic and archaeal chaperonins. These interior surface chemical properties likely play an important role in TRiC's cellular substrate specificity.
Conflict of interest statement
Conflict of interest statement: The Sponsor is an investigator of the Nanomedicince Development Center with the support of National Institutes of Health Grant (5PN2EY016525), which is directed by the corresponding author.
Figures



References
-
- Scott MD, Frydman J. Aberrant protein folding as the molecular basis of cancer. Method Mol Biol. 2003;232:67–76. - PubMed
-
- Dobson CM. Principles of protein folding, misfolding and aggregation. Semin Cell Dev Biol. 2004;15:3–16. - PubMed
-
- Frydman J. Folding of newly translated proteins in vivo: The role of molecular chaperones. Annu Rev Biochem. 2001;70:603–647. - PubMed
-
- Hartl FU, Hayer-Hartl M. Converging concepts of protein folding in vitro and in vivo. Nat Struct Mol Biol. 2009;16:574–581. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

