Structural basis for substrate selectivity in human maltase-glucoamylase and sucrase-isomaltase N-terminal domains
- PMID: 20356844
- PMCID: PMC2878540
- DOI: 10.1074/jbc.M109.078980
Structural basis for substrate selectivity in human maltase-glucoamylase and sucrase-isomaltase N-terminal domains
Abstract
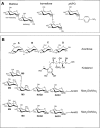
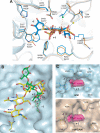
Human maltase-glucoamylase (MGAM) and sucrase-isomaltase (SI) are small intestinal enzymes that work concurrently to hydrolyze the mixture of linear alpha-1,4- and branched alpha-1,6-oligosaccharide substrates that typically make up terminal starch digestion products. MGAM and SI are each composed of duplicated catalytic domains, N- and C-terminal, which display overlapping substrate specificities. The N-terminal catalytic domain of human MGAM (ntMGAM) has a preference for short linear alpha-1,4-oligosaccharides, whereas N-terminal SI (ntSI) has a broader specificity for both alpha-1,4- and alpha-1,6-oligosaccharides. Here we present the crystal structure of the human ntSI, in apo form to 3.2 A and in complex with the inhibitor kotalanol to 2.15 A resolution. Structural comparison with the previously solved structure of ntMGAM reveals key active site differences in ntSI, including a narrow hydrophobic +1 subsite, which may account for its additional substrate specificity for alpha-1,6 substrates.
Figures

References
-
- Van Beers E. H., Büller H. A., Grand R. J., Einerhand A. W., Dekker J. (1995) Crit. Rev. Biochem. Mol. Biol. 30, 197–262 - PubMed
-
- Semenza G. (1986) Ann. Rev. Cell Biol. 2, 255–313 - PubMed
-
- Quezada-Calvillo R., Sim L., Ao Z., Hamaker B. R., Quaroni A., Brayer G. D., Sterchi E. E., Robayo-Torres C. C., Rose D. R., Nichols B. L. (2008) J. Nutr. 138, 685–692 - PubMed
-
- Heymann H., Breitmeier D., Günther S. (1995) Biol. Chem. Hoppe Seyler 376, 249–253 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

