Molecular Insights into Inhibition of the Methylated Histone-Plant Homeodomain Complexes by Calixarenes
- PMID: 26229108
- PMCID: PMC4645621
- DOI: 10.1074/jbc.M115.669333
Molecular Insights into Inhibition of the Methylated Histone-Plant Homeodomain Complexes by Calixarenes
Abstract
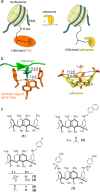
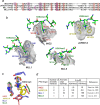
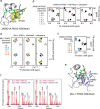
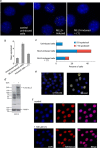
Plant homeodomain (PHD) finger-containing proteins are implicated in fundamental biological processes, including transcriptional activation and repression, DNA damage repair, cell differentiation, and survival. The PHD finger functions as an epigenetic reader that binds to posttranslationally modified or unmodified histone H3 tails, recruiting catalytic writers and erasers and other components of the epigenetic machinery to chromatin. Despite the critical role of the histone-PHD interaction in normal and pathological processes, selective inhibitors of this association have not been well developed. Here we demonstrate that macrocyclic calixarenes can disrupt binding of PHD fingers to methylated lysine 4 of histone H3 in vitro and in vivo. The inhibitory activity relies on differences in binding affinities of the PHD fingers for H3K4me and the methylation state of the histone ligand, whereas the composition of the aromatic H3K4me-binding site of the PHD fingers appears to have no effect. Our approach provides a novel tool for studying the biological roles of methyllysine readers in epigenetic signaling.
Keywords: PHD finger; calixarene; chromatin; histone; histone methylation; inhibitor.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




References
-
- Shi X., Hong T., Walter K. L., Ewalt M., Michishita E., Hung T., Carney D., Peña P., Lan F., Kaadige M. R., Lacoste N., Cayrou C., Davrazou F., Saha A., Cairns B. R., Ayer D. E., Kutateladze T. G., Shi Y., Côté J., Chua K. F., Gozani O. (2006) ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 442, 96–99 - PMC - PubMed
-
- Wysocka J., Swigut T., Xiao H., Milne T. A., Kwon S. Y., Landry J., Kauer M., Tackett A. J., Chait B. T., Badenhorst P., Wu C., Allis C. D. (2006) A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442, 86–90 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

