Structural basis for activation of class Ib ribonucleotide reductase
- PMID: 20688982
- PMCID: PMC3020666
- DOI: 10.1126/science.1190187
Structural basis for activation of class Ib ribonucleotide reductase
Abstract
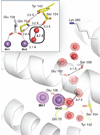
The class Ib ribonucleotide reductase of Escherichia coli can initiate reduction of nucleotides to deoxynucleotides with either a Mn(III)2-tyrosyl radical (Y•) or a Fe(III)2-Y• cofactor in the NrdF subunit. Whereas Fe(III)2-Y• can self-assemble from Fe(II)2-NrdF and O2, activation of Mn(II)2-NrdF requires a reduced flavoprotein, NrdI, proposed to form the oxidant for cofactor assembly by reduction of O2. The crystal structures reported here of E. coli Mn(II)2-NrdF and Fe(II)2-NrdF reveal different coordination environments, suggesting distinct initial binding sites for the oxidants during cofactor activation. In the structures of Mn(II)2-NrdF in complex with reduced and oxidized NrdI, a continuous channel connects the NrdI flavin cofactor to the NrdF Mn(II)2 active site. Crystallographic detection of a putative peroxide in this channel supports the proposed mechanism of Mn(III)2-Y• cofactor assembly.
Figures



Comment in
-
Biochemistry. A never-ending story.Science. 2010 Sep 17;329(5998):1475-6. doi: 10.1126/science.1196347. Science. 2010. PMID: 20847256 No abstract available.
References
-
- Nordlund P, Reichard P. Annu. Rev. Biochem. 2006;75:681. - PubMed
-
- Gon S, Faulkner MJ, Beckwith J. Antioxid. Redox Signal. 2006;8:735. - PubMed
-
- McHugh JP, et al. J. Biol. Chem. 2003;278:29478. - PubMed
-
- Monje-Casas F, Jurado J, Prieto-Alamo MJ, Holmgren A, Pueyo C. J. Biol. Chem. 2001;276:18031. - PubMed
-
- Atkin CL, Thelander L, Reichard P, Lang G. J. Biol. Chem. 1973;248:7464. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

