An unusual dimeric structure and assembly for TLR4 regulator RP105-MD-1
- PMID: 21857663
- PMCID: PMC3362203
- DOI: 10.1038/nsmb.2106
An unusual dimeric structure and assembly for TLR4 regulator RP105-MD-1
Abstract
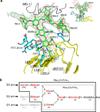
RP105-MD-1 modulates the TLR4-MD-2-mediated, innate immune response against bacterial lipopolysaccharide (LPS). The crystal structure of the bovine 1:1 RP105-MD-1 complex bound to a putative endogenous lipid at 2.9 Å resolution shares a similar overall architecture to its homolog TLR4-MD-2 but assembles into an unusual 2:2 homodimer that differs from any other known TLR-ligand assembly. The homodimer is assembled in a head-to-head orientation that juxtaposes the N-terminal leucine-rich repeats (LRRs) of the two RP105 chains, rather than the usual tail-to-tail configuration of C-terminal LRRs in ligand-activated TLR dimers, such as TLR1-TRL2, TLR2-TLR6, TLR3-TLR3 and TLR4-TLR4. Another unusual interaction is mediated by an RP105-specific asparagine-linked glycan, which wedges MD-1 into the co-receptor binding concavity on RP105. This unique mode of assembly represents a new paradigm for TLR complexes and suggests a molecular mechanism for regulating LPS responses.
Figures

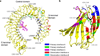



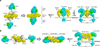
References
-
- Bell JK, et al. Leucine-rich repeats and pathogen recognition in Toll-like receptors. Trends Immunol. 2003;24:528–533. - PubMed
-
- Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. 2003;21:335–376. - PubMed
-
- Nakhaei P, Genin P, Civas A, Hiscott J. RIG-I-like receptors: sensing and responding to RNA virus infection. Semin Immunol. 2009;21:215–222. - PubMed
-
- Kim HM, et al. Crystal structure of the TLR4-MD-2 complex with bound endotoxin antagonist Eritoran. Cell. 2007;130:906–917. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

