Structures of Proline Utilization A (PutA) Reveal the Fold and Functions of the Aldehyde Dehydrogenase Superfamily Domain of Unknown Function
- PMID: 27679491
- PMCID: PMC5104932
- DOI: 10.1074/jbc.M116.756965
Structures of Proline Utilization A (PutA) Reveal the Fold and Functions of the Aldehyde Dehydrogenase Superfamily Domain of Unknown Function
Abstract
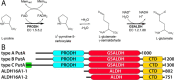
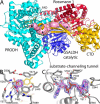
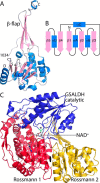
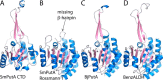
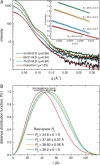
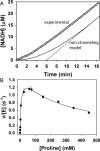
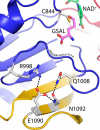
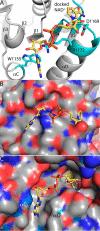
Aldehyde dehydrogenases (ALDHs) catalyze the NAD(P)+-dependent oxidation of aldehydes to carboxylic acids and are important for metabolism and detoxification. Although the ALDH superfamily fold is well established, some ALDHs contain an uncharacterized domain of unknown function (DUF) near the C terminus of the polypeptide chain. Herein, we report the first structure of a protein containing the ALDH superfamily DUF. Proline utilization A from Sinorhizobium meliloti (SmPutA) is a 1233-residue bifunctional enzyme that contains the DUF in addition to proline dehydrogenase and l-glutamate-γ-semialdehyde dehydrogenase catalytic modules. Structures of SmPutA with a proline analog bound to the proline dehydrogenase site and NAD+ bound to the ALDH site were determined in two space groups at 1.7-1.9 Å resolution. The DUF consists of a Rossmann dinucleotide-binding fold fused to a three-stranded β-flap. The Rossmann domain resembles the classic ALDH superfamily NAD+-binding domain, whereas the flap is strikingly similar to the ALDH superfamily dimerization domain. Paradoxically, neither structural element performs its implied function. Electron density maps show that NAD+ does not bind to the DUF Rossmann fold, and small-angle X-ray scattering reveals a novel dimer that has never been seen in the ALDH superfamily. The structure suggests that the DUF is an adapter domain that stabilizes the aldehyde substrate binding loop and seals the substrate-channeling tunnel via tertiary structural interactions that mimic the quaternary structural interactions found in non-DUF PutAs. Kinetic data for SmPutA indicate a substrate-channeling mechanism, in agreement with previous studies of other PutAs.
Keywords: X-ray crystallography; aldehyde dehydrogenase superfamily; enzyme kinetics; flavoprotein; nicotinamide adenine dinucleotide (NAD); oligomerization; proline catabolism; proline utilization A; protein domain; small-angle X-ray scattering (SAXS).
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

References
-
- Liu Z. J., Sun Y. J., Rose J., Chung Y. J., Hsiao C. D., Chang W. R., Kuo I., Perozich J., Lindahl R., Hempel J., and Wang B. C. (1997) The first structure of an aldehyde dehydrogenase reveals novel interactions between NAD and the Rossmann fold. Nat. Struct. Biol. 4, 317–326 - PubMed
-
- Tanner J. J., and Becker D. F. (2013) PutA and proline metabolism. In Handbook of Flavoproteins: Volume 1. Oxidases, Dehydrogenases and Related Systems (Hille R., Miller S. M., and Palfey B. A., eds) pp. 31–56, De Gruyter, Berlin
-
- Srivastava D., Schuermann J. P., White T. A., Krishnan N., Sanyal N., Hura G. L., Tan A., Henzl M. T., Becker D. F., and Tanner J. J. (2010) Crystal structure of the bifunctional proline utilization A flavoenzyme from Bradyrhizobium japonicum. Proc. Natl. Acad. Sci. U.S.A. 107, 2878–2883 - PMC - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

