Structural basis for clonal diversity of the human T-cell response to a dominant influenza virus epitope
- PMID: 28931605
- PMCID: PMC5683187
- DOI: 10.1074/jbc.M117.810382
Structural basis for clonal diversity of the human T-cell response to a dominant influenza virus epitope
Abstract
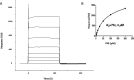
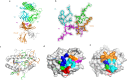
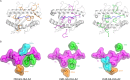
Influenza A virus (IAV) causes an acute infection in humans that is normally eliminated by CD8+ cytotoxic T lymphocytes. Individuals expressing the MHC class I molecule HLA-A2 produce cytotoxic T lymphocytes bearing T-cell receptors (TCRs) that recognize the immunodominant IAV epitope GILGFVFTL (GIL). Most GIL-specific TCRs utilize α/β chain pairs encoded by the TRAV27/TRBV19 gene combination to recognize this relatively featureless peptide epitope (canonical TCRs). However, ∼40% of GIL-specific TCRs express a wide variety of other TRAV/TRBV combinations (non-canonical TCRs). To investigate the structural underpinnings of this remarkable diversity, we determined the crystal structure of a non-canonical GIL-specific TCR (F50) expressing the TRAV13-1/TRBV27 gene combination bound to GIL-HLA-A2 to 1.7 Å resolution. Comparison of the F50-GIL-HLA-A2 complex with the previously published complex formed by a canonical TCR (JM22) revealed that F50 and JM22 engage GIL-HLA-A2 in markedly different orientations. These orientations are distinguished by crossing angles of TCR to peptide-MHC of 29° for F50 versus 69° for JM22 and by a focus by F50 on the C terminus rather than the center of the MHC α1 helix for JM22. In addition, F50, unlike JM22, uses a tryptophan instead of an arginine to fill a critical notch between GIL and the HLA-A2 α2 helix. The F50-GIL-HLA-A2 complex shows that there are multiple structurally distinct solutions to recognizing an identical peptide-MHC ligand with sufficient affinity to elicit a broad anti-IAV response that protects against viral escape and T-cell clonal loss.
Keywords: T-cell receptor; crystal structure; epitope; influenza virus; major histocompatibility complex (MHC); protein complex; surface plasmon resonance (SPR).
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures
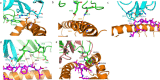
Similar articles
-
The immunodominant influenza A virus M158-66 cytotoxic T lymphocyte epitope exhibits degenerate class I major histocompatibility complex restriction in humans.J Virol. 2014 Sep;88(18):10613-23. doi: 10.1128/JVI.00855-14. Epub 2014 Jul 2. J Virol. 2014. PMID: 24990997 Free PMC article.
-
Tax and M1 peptide/HLA-A2-specific Fabs and T cell receptors recognize nonidentical structural features on peptide/HLA-A2 complexes.J Immunol. 2003 Sep 15;171(6):3064-74. doi: 10.4049/jimmunol.171.6.3064. J Immunol. 2003. PMID: 12960332
-
Diversity and dominance among TCR recognizing HLA-A2.1+ influenza matrix peptide in human MHC class I transgenic mice.J Immunol. 1994 Nov 15;153(10):4458-67. J Immunol. 1994. PMID: 7963521
-
T cells and their eons-old obsession with MHC.Immunol Rev. 2012 Nov;250(1):49-60. doi: 10.1111/imr.12004. Immunol Rev. 2012. PMID: 23046122 Free PMC article. Review.
-
Epitope prediction and identification- adaptive T cell responses in humans.Semin Immunol. 2020 Aug;50:101418. doi: 10.1016/j.smim.2020.101418. Epub 2020 Oct 31. Semin Immunol. 2020. PMID: 33131981 Free PMC article. Review.
Cited by
-
GIANA allows computationally-efficient TCR clustering and multi-disease repertoire classification by isometric transformation.Nat Commun. 2021 Aug 4;12(1):4699. doi: 10.1038/s41467-021-25006-7. Nat Commun. 2021. PMID: 34349111 Free PMC article.
-
Characterization of amino acid residues of T-cell receptors interacting with HLA-A*02-restricted antigen peptides.Ann Transl Med. 2021 Mar;9(6):495. doi: 10.21037/atm-21-835. Ann Transl Med. 2021. PMID: 33850892 Free PMC article.
-
Predicting T Cell Receptor Antigen Specificity From Structural Features Derived From Homology Models of Receptor-Peptide-Major Histocompatibility Complexes.Front Physiol. 2021 Sep 8;12:730908. doi: 10.3389/fphys.2021.730908. eCollection 2021. Front Physiol. 2021. PMID: 34566692 Free PMC article.
-
Immunogenic SARS-CoV-2 Epitopes: In Silico Study Towards Better Understanding of COVID-19 Disease-Paving the Way for Vaccine Development.Vaccines (Basel). 2020 Jul 23;8(3):408. doi: 10.3390/vaccines8030408. Vaccines (Basel). 2020. PMID: 32717854 Free PMC article.
-
Different T-cell and B-cell repertoire elicited by the SARS-CoV-2 inactivated vaccine and S1 subunit vaccine in rhesus macaques.Hum Vaccin Immunother. 2022 Nov 30;18(6):2118477. doi: 10.1080/21645515.2022.2118477. Epub 2022 Sep 7. Hum Vaccin Immunother. 2022. PMID: 36070519 Free PMC article.
References
-
- Arstila T. P., Casrouge A., Baron V., Even J., Kanellopoulos J., and Kourilsky P. (1999) A direct estimate of the human αβ T cell receptor diversity. Science 286, 958–961 - PubMed
-
- Li H. M., Hiroi Z., Y., Shi A., Chen G., De S., Metter E. J., Wood W. H. 3rd, Sharov A., Milner J. D., Becker K. G., Zhan M., and Weng N. P. (2016) TCRβ repertoire of CD4+ and CD8+ T cells is distinct in richness, distribution, and CDR3 amino acid composition. J. Leukoc. Biol. 99, 505–513 - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

