Molecular mechanism of strict substrate specificity of an extradiol dioxygenase, DesB, derived from Sphingobium sp. SYK-6
- PMID: 24657997
- PMCID: PMC3962378
- DOI: 10.1371/journal.pone.0092249
Molecular mechanism of strict substrate specificity of an extradiol dioxygenase, DesB, derived from Sphingobium sp. SYK-6
Abstract
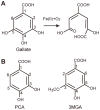
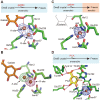
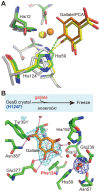
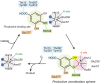
DesB, which is derived from Sphingobium sp. SYK-6, is a type II extradiol dioxygenase that catalyzes a ring opening reaction of gallate. While typical extradiol dioxygenases show broad substrate specificity, DesB has strict substrate specificity for gallate. The substrate specificity of DesB seems to be required for the efficient growth of S. sp. SYK-6 using lignin-derived aromatic compounds. Since direct coordination of hydroxyl groups of the substrate to the non-heme iron in the active site is a critical step for the catalytic reaction of the extradiol dioxygenases, the mechanism of the substrate recognition and coordination of DesB was analyzed by biochemical and crystallographic methods. Our study demonstrated that the direct coordination between the non-heme iron and hydroxyl groups of the substrate requires a large shift of the Fe (II) ion in the active site. Mutational analysis revealed that His124 and His192 in the active site are essential to the catalytic reaction of DesB. His124, which interacts with OH (4) of the bound gallate, seems to contribute to proper positioning of the substrate in the active site. His192, which is located close to OH (3) of the gallate, is likely to serve as the catalytic base. Glu377' interacts with OH (5) of the gallate and seems to play a critical role in the substrate specificity. Our biochemical and structural study showed the substrate recognition and catalytic mechanisms of DesB.
Conflict of interest statement
Figures
References
-
- Sterner R, Liebl W (2001) Thermophilic adaptation of proteins. Crit Rev Biochem Mol Biol 36: 39–106. - PubMed
-
- Katayama Y, Nishikawa S, Murayama A, Yamasaki M, Morohoshi N, et al. (1988) The metabolism of biphenyl structures in lignin by the soil bacterium (Pseudomonas paucimobilis SYK-6). FEBS Lett 233: 129–133. - PubMed
-
- Masai E, Katayama Y, Fukuda M (2007) Genetic and biochemical investigations on bacterial catabolic pathways for lignin-derived aromatic compounds. Biosci Biotechnol Biochem 71: 1–15. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

