The class III cyclobutane pyrimidine dimer photolyase structure reveals a new antenna chromophore binding site and alternative photoreduction pathways
- PMID: 25784552
- PMCID: PMC4416854
- DOI: 10.1074/jbc.M115.637868
The class III cyclobutane pyrimidine dimer photolyase structure reveals a new antenna chromophore binding site and alternative photoreduction pathways
Abstract
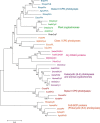
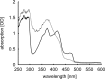
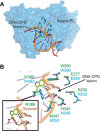
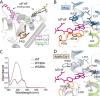
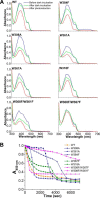
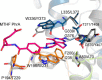
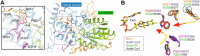
Photolyases are proteins with an FAD chromophore that repair UV-induced pyrimidine dimers on the DNA in a light-dependent manner. The cyclobutane pyrimidine dimer class III photolyases are structurally unknown but closely related to plant cryptochromes, which serve as blue-light photoreceptors. Here we present the crystal structure of a class III photolyase termed photolyase-related protein A (PhrA) of Agrobacterium tumefaciens at 1.67-Å resolution. PhrA contains 5,10-methenyltetrahydrofolate (MTHF) as an antenna chromophore with a unique binding site and mode. Two Trp residues play pivotal roles for stabilizing MTHF by a double π-stacking sandwich. Plant cryptochrome I forms a pocket at the same site that could accommodate MTHF or a similar molecule. The PhrA structure and mutant studies showed that electrons flow during FAD photoreduction proceeds via two Trp triads. The structural studies on PhrA give a clearer picture on the evolutionary transition from photolyase to photoreceptor.
Keywords: Cryptochrome; Crystal Structure; DNA Repair; Flavin Adenine Dinucleotide (FAD); Molecular Evolution; Mutagenesis in Vitro; Photobiology; Photoreceptor; Photoreduction; Site-directed Mutagenesis.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


References
-
- Sancar A. (2003) Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem. Rev. 103, 2203–2237 - PubMed
-
- Müller M., Carell T. (2009) Structural biology of DNA photolyases and cryptochromes. Curr. Opin. Struct. Biol. 19, 277–285 - PubMed
-
- Chaves I., Pokorny R., Byrdin M., Hoang N., Ritz T., Brettel K., Essen L. O., van der Horst G. T., Batschauer A., Ahmad M. (2011) The cryptochromes: blue light photoreceptors in plants and animals. Annu. Rev. Plant Biol. 62, 335–364 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

