Csy4 relies on an unusual catalytic dyad to position and cleave CRISPR RNA
- PMID: 22522703
- PMCID: PMC3380207
- DOI: 10.1038/emboj.2012.107
Csy4 relies on an unusual catalytic dyad to position and cleave CRISPR RNA
Abstract
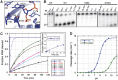
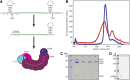
CRISPR-Cas adaptive immune systems protect prokaryotes against foreign genetic elements. crRNAs derived from CRISPR loci base pair with complementary nucleic acids, leading to their destruction. In Pseudomonas aeruginosa, crRNA biogenesis requires the endoribonuclease Csy4, which binds and cleaves the repetitive sequence of the CRISPR transcript. Biochemical assays and three co-crystal structures of wild-type and mutant Csy4/RNA complexes reveal a substrate positioning and cleavage mechanism in which a histidine deprotonates the ribosyl 2'-hydroxyl pinned in place by a serine, leading to nucleophilic attack on the scissile phosphate. The active site catalytic dyad lacks a general acid to protonate the leaving group and positively charged residues to stabilize the transition state, explaining why the observed catalytic rate constant is ∼10(4)-fold slower than that of RNase A. We show that this RNA cleavage step is essential for assembly of the Csy protein-crRNA complex that facilitates target recognition. Considering that Csy4 recognizes a single cellular substrate and sequesters the cleavage product, evolutionary pressure has likely selected for substrate specificity and high-affinity crRNA interactions at the expense of rapid cleavage kinetics.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
-
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 - PMC - PubMed
-
- Al-Attar S, Westra ER, van der Oost J, Brouns SJ (2011) Clustered regularly interspaced short palindromic repeats (CRISPRs): the hallmark of an ingenious antiviral defense mechanism in prokaryotes. Biol Chem 392: 277–289 - PubMed
-
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315: 1709–1712 - PubMed
-
- Bolotin A, Ouinquis B, Sorokin A, Ehrlich SD (2005) Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology-Sgm 151: 2551–2561 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

