4'-O-substitutions determine selectivity of aminoglycoside antibiotics
- PMID: 24473108
- PMCID: PMC3942853
- DOI: 10.1038/ncomms4112
4'-O-substitutions determine selectivity of aminoglycoside antibiotics
Abstract
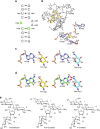
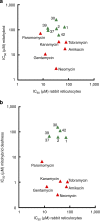
Clinical use of 2-deoxystreptamine aminoglycoside antibiotics, which target the bacterial ribosome, is compromised by adverse effects related to limited drug selectivity. Here we present a series of 4',6'-O-acetal and 4'-O-ether modifications on glucopyranosyl ring I of aminoglycosides. Chemical modifications were guided by measuring interactions between the compounds synthesized and ribosomes harbouring single point mutations in the drug-binding site, resulting in aminoglycosides that interact poorly with the drug-binding pocket of eukaryotic mitochondrial or cytosolic ribosomes. Yet, these compounds largely retain their inhibitory activity for bacterial ribosomes and show antibacterial activity. Our data indicate that 4'-O-substituted aminoglycosides possess increased selectivity towards bacterial ribosomes and little activity for any of the human drug-binding pockets.
Conflict of interest statement
E.C.B, D.P.-F. and A.V. are co-inventors on a patent filed by the University of Zurich on 4′ modifications of disubstituted 2-deoxystreptamines (WO2008/092690A10). The remaining authors declare no competing financial interests. The University of Zurich has filed a patent application on 4′ modifications of disubstituted 2-deoxystreptamines (WO2008/092690A1).
Figures

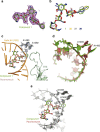

References
-
- Poehlsgaard J. & Douthwaite S. The bacterial ribosome as a target for antibiotics. Nat. Rev. Microbiol. 3, 870–881 (2005). - PubMed
-
- Wilson D. N. The A-Z of bacterial translation inhibitors. Crit. Rev. Biochem. Mol. Biol. 44, 393–433 (2009). - PubMed
-
- World Health Organisation, Report of the second WHO Expert Meeting, Copenhagen, Denmark (2007) Critically important antimicrobials for human health, http://www.who.int/foodborne_disease/resistance/antimicrobials_human.pdf (WHO, Geneva, Switzerland).
-
- Chambers H. F. Chemotherapy of microbial diseases. In: Goodmann & Gilman’sThe Pharmaceutical Basis of Therapeutics 10th edn Eds Hardman J. G., Limbird L. E. 1103–1121The McGraw-Hill Companies (1996).
-
- Forge A. & Schacht J. Aminoglycoside antibiotics. Audiol. Neurootol. 5, 3–22 (2000). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

