High-resolution structures of the IgM Fc domains reveal principles of its hexamer formation
- PMID: 23733956
- PMCID: PMC3690842
- DOI: 10.1073/pnas.1300547110
High-resolution structures of the IgM Fc domains reveal principles of its hexamer formation
Abstract
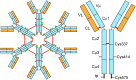
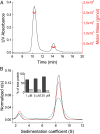
IgM is the first antibody produced during the humoral immune response. Despite its fundamental role in the immune system, IgM is structurally only poorly described. In this work we used X-ray crystallography and NMR spectroscopy to determine the atomic structures of the constant IgM Fc domains (Cµ2, Cµ3, and Cµ4) and to address their roles in IgM oligomerization. Although the isolated domains share the typical Ig fold, they differ substantially in dimerization properties and quaternary contacts. Unexpectedly, the Cµ4 domain and its C-terminal tail piece are responsible and sufficient for the specific polymerization of Cµ4 dimers into covalently linked hexamers of dimers. Based on small angle X-ray scattering data, we present a model of the ring-shaped Cµ4 structure, which reveals the principles of IgM oligomerization.
Keywords: antibody oligomerization; dimer interfaces; hybrid approach.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Wannemuehler MJ, Galvin JE. Bacterial immunogens and protective immunity in swine. Vet Immunol Immunopathol. 1994;43(1-3):117–126. - PubMed
-
- Ohanian SH, Schlager SI. Humoral immune killing of nucleated cells: mechanisms of complement-mediated attack and target cell defense. Crit Rev Immunol. 1981;1(3):165–209. - PubMed
-
- Boes M. Role of natural and immune IgM antibodies in immune responses. Mol Immunol. 2000;37(18):1141–1149. - PubMed
-
- Taylor B, et al. C1q binding properties of monomer and polymer forms of mouse IgM mu-chain variants. Pro544Gly and Pro434Ala. J Immunol. 1994;153(11):5303–5313. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

