Structure of the cyanobacterial phytochrome 2 photosensor implies a tryptophan switch for phytochrome signaling
- PMID: 24174528
- PMCID: PMC3861623
- DOI: 10.1074/jbc.M113.510461
Structure of the cyanobacterial phytochrome 2 photosensor implies a tryptophan switch for phytochrome signaling
Abstract
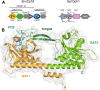
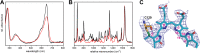
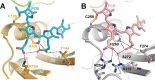
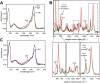
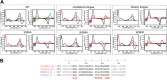
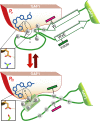
Phytochromes are highly versatile photoreceptors, which occur ubiquitously in plants as well as in many light-responsive microorganisms. Here, photosynthetic cyanobacteria utilize up to three different phytochrome architectures, where only the plant-like and the single-domain cyanobacteriochromes are structurally characterized so far. Cph2 represents a third group in Synechocystis species and affects their capability of phototaxis by controlling c-di-GMP synthesis and degradation. The 2.6-Å crystal structure of its red/far-red responsive photosensory module in the Pr state reveals a tandem-GAF bidomain that lacks the figure-of-eight knot of the plant/cph1 subfamily. Its covalently attached phycocyanobilin chromophore adopts a highly tilted ZZZssa conformation with a novel set of interactions between its propionates and the GAF1 domain. The tongue-like protrusion from the GAF2 domain interacts with the GAF1-bound chromophore via its conserved PRXSF, WXE, and W(G/A)G motifs. Mutagenesis showed that the integrity of the tongue is indispensable for Pr → Pfr photoconversion and involves a swap of the motifs' tryptophans within the tongue-GAF1 interface. This "Trp switch" is supposed to be a crucial element for the photochromicity of all multidomain phytochromes.
Keywords: Biliprotein; Cyanobacteria; Photochromicity; Phytochrome; Protein Conformation; Red Light Photoreceptor; Signal Transduction; Signaling; Structural Biology; c-di-GMP Signaling.
Figures



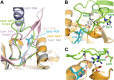
References
-
- Hughes J., Lamparter T., Mittmann F., Hartmann E., Gärtner W., Wilde A., Börner T. (1997) A prokaryotic phytochrome. Nature 386, 663. - PubMed
-
- Rodriguez-Romero J., Hedtke M., Kastner C., Müller S., Fischer R. (2010) Fungi, hidden in soil or up in the air. Light makes a difference. Annu. Rev. Microbiol. 64, 585–610 - PubMed
-
- Nagatani A. (2010) Phytochrome. Structural basis for its functions. Curr. Opin. Plant Biol. 13, 565–570 - PubMed
-
- Vierstra R. D., Zhang J. (2011) Phytochrome signaling. Solving the Gordian knot with microbial relatives. Trends Plant Sci. 16, 417–426 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

