Structural basis of the regulatory mechanism of the plant CIPK family of protein kinases controlling ion homeostasis and abiotic stress
- PMID: 25288725
- PMCID: PMC4210280
- DOI: 10.1073/pnas.1407610111
Structural basis of the regulatory mechanism of the plant CIPK family of protein kinases controlling ion homeostasis and abiotic stress
Abstract
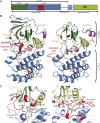
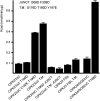
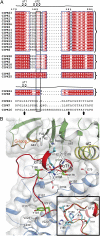
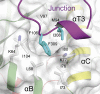
Plant cells have developed specific protective molecular machinery against environmental stresses. The family of CBL-interacting protein kinases (CIPK) and their interacting activators, the calcium sensors calcineurin B-like (CBLs), work together to decode calcium signals elicited by stress situations. The molecular basis of biological activation of CIPKs relies on the calcium-dependent interaction of a self-inhibitory NAF motif with a particular CBL, the phosphorylation of the activation loop by upstream kinases, and the subsequent phosphorylation of the CBL by the CIPK. We present the crystal structures of the NAF-truncated and pseudophosphorylated kinase domains of CIPK23 and CIPK24/SOS2. In addition, we provide biochemical data showing that although CIPK23 is intrinsically inactive and requires an external stimulation, CIPK24/SOS2 displays basal activity. This data correlates well with the observed conformation of the respective activation loops: Although the loop of CIPK23 is folded into a well-ordered structure that blocks the active site access to substrates, the loop of CIPK24/SOS2 protrudes out of the active site and allows catalysis. These structures together with biochemical and biophysical data show that CIPK kinase activity necessarily requires the coordinated releases of the activation loop from the active site and of the NAF motif from the nucleotide-binding site. Taken all together, we postulate the basis for a conserved calcium-dependent NAF-mediated regulation of CIPKs and a variable regulation by upstream kinases.
Keywords: abiotic stress; ion transport; signaling.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Batistič O, Kudla J. Analysis of calcium signaling pathways in plants. Biochim Biophys Acta. 2012;1820(8):1283–1293. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

