Structure, activity, and inhibition of the Carboxyltransferase β-subunit of acetyl coenzyme A carboxylase (AccD6) from Mycobacterium tuberculosis
- PMID: 25092705
- PMCID: PMC4187906
- DOI: 10.1128/AAC.02574-13
Structure, activity, and inhibition of the Carboxyltransferase β-subunit of acetyl coenzyme A carboxylase (AccD6) from Mycobacterium tuberculosis
Abstract
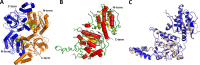
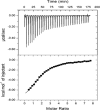
In Mycobacterium tuberculosis, the carboxylation of acetyl coenzyme A (acetyl-CoA) to produce malonyl-CoA, a building block in long-chain fatty acid biosynthesis, is catalyzed by two enzymes working sequentially: a biotin carboxylase (AccA) and a carboxyltransferase (AccD). While the exact roles of the three different biotin carboxylases (AccA1 to -3) and the six carboxyltransferases (AccD1 to -6) in M. tuberculosis are still not clear, AccD6 in complex with AccA3 can synthesize malonyl-CoA from acetyl-CoA. A series of 10 herbicides that target plant acetyl-CoA carboxylases (ACC) were tested for inhibition of AccD6 and for whole-cell activity against M. tuberculosis. From the tested herbicides, haloxyfop, an arylophenoxypropionate, showed in vitro inhibition of M. tuberculosis AccD6, with a 50% inhibitory concentration (IC50) of 21.4 ± 1 μM. Here, we report the crystal structures of M. tuberculosis AccD6 in the apo form (3.0 Å) and in complex with haloxyfop-R (2.3 Å). The structure of M. tuberculosis AccD6 in complex with haloxyfop-R shows two molecules of the inhibitor bound on each AccD6 subunit. These results indicate the potential for developing novel therapeutics for tuberculosis based on herbicides with low human toxicity.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures



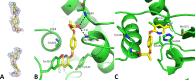



References
-
- Tanabe T, Wada K, Okazaki T, Numa S. 1975. Acetyl-coenzyme-A carboxylase from rat liver. Subunit structure and proteolytic modification. Eur. J. Biochem. 57:15–24 - PubMed
-
- Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Conner R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544. 10.1038/31159 - DOI - PubMed
-
- Lin TW, Melgar MM, Kurth D, Swamidass SJ, Purdon J, Tseng T, Gago G, Baldi P, Gramajo H, Tsai SC. 2006. Structure-based inhibitor design of AccD5, an essential acyl-CoA carboxylase carboxyltransferase domain of Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 103:3072–3077. 10.1073/pnas.0510580103 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

