Structural characterization of a unique marine animal family 7 cellobiohydrolase suggests a mechanism of cellulase salt tolerance
- PMID: 23733951
- PMCID: PMC3690837
- DOI: 10.1073/pnas.1301502110
Structural characterization of a unique marine animal family 7 cellobiohydrolase suggests a mechanism of cellulase salt tolerance
Abstract
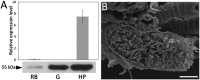
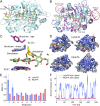
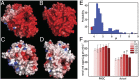
Nature uses a diversity of glycoside hydrolase (GH) enzymes to convert polysaccharides to sugars. As lignocellulosic biomass deconstruction for biofuel production remains costly, natural GH diversity offers a starting point for developing industrial enzymes, and fungal GH family 7 (GH7) cellobiohydrolases, in particular, provide significant hydrolytic potential in industrial mixtures. Recently, GH7 enzymes have been found in other kingdoms of life besides fungi, including in animals and protists. Here, we describe the in vivo spatial expression distribution, properties, and structure of a unique endogenous GH7 cellulase from an animal, the marine wood borer Limnoria quadripunctata (LqCel7B). RT-quantitative PCR and Western blot studies show that LqCel7B is expressed in the hepatopancreas and secreted into the gut for wood degradation. We produced recombinant LqCel7B, with which we demonstrate that LqCel7B is a cellobiohydrolase and obtained four high-resolution crystal structures. Based on a crystallographic and computational comparison of LqCel7B to the well-characterized Hypocrea jecorina GH7 cellobiohydrolase, LqCel7B exhibits an extended substrate-binding motif at the tunnel entrance, which may aid in substrate acquisition and processivity. Interestingly, LqCel7B exhibits striking surface charges relative to fungal GH7 enzymes, which likely results from evolution in marine environments. We demonstrate that LqCel7B stability and activity remain unchanged, or increase at high salt concentration, and that the L. quadripunctata GH mixture generally contains cellulolytic enzymes with highly acidic surface charge compared with enzymes derived from terrestrial microbes. Overall, this study suggests that marine cellulases offer significant potential for utilization in high-solids industrial biomass conversion processes.
Keywords: carbohydrate degrading enzymes; gribble.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Somerville C. Cellulose synthesis in higher plants. Annu Rev Cell Dev Biol. 2006;22:53–78. - PubMed
-
- Himmel ME, et al. Biomass recalcitrance: Engineering plants and enzymes for biofuels production. Science. 2007;315(5813):804–807. - PubMed
-
- Carroll A, Somerville C. Cellulosic biofuels. Annu Rev Plant Biol. 2009;60:165–182. - PubMed
-
- Burton RA, Gidley MJ, Fincher GB. Heterogeneity in the chemistry, structure and function of plant cell walls. Nat Chem Biol. 2010;6(10):724–732. - PubMed
-
- Chundawat SPS, Beckham GT, Himmel ME, Dale BE. Deconstruction of lignocellulosic biomass to fuels and chemicals. Annu Rev Chem Biomol Eng. 2011;2:121–145. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

