Structural basis for recruitment and activation of the AP-1 clathrin adaptor complex by Arf1
- PMID: 23415225
- PMCID: PMC3913725
- DOI: 10.1016/j.cell.2012.12.042
Structural basis for recruitment and activation of the AP-1 clathrin adaptor complex by Arf1
Abstract
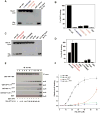
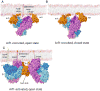
AP-1 is a clathrin adaptor complex that sorts cargo between the trans-Golgi network and endosomes. AP-1 recruitment to these compartments requires Arf1-GTP. The crystal structure of the tetrameric core of AP-1 in complex with Arf1-GTP, together with biochemical analyses, shows that Arf1 activates cargo binding by unlocking AP-1. Unlocking is driven by two molecules of Arf1 that bridge two copies of AP-1 at two interaction sites. The GTP-dependent switch I and II regions of Arf1 bind to the N terminus of the β1 subunit of one AP-1 complex, while the back side of Arf1 binds to the central part of the γ subunit trunk of a second AP-1 complex. A third Arf1 interaction site near the N terminus of the γ subunit is important for recruitment, but not activation. These observations lead to a model for the recruitment and activation of AP-1 by Arf1.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures





References
-
- Amor JC, Harrison DH, Kahn RA, Ringe D. Structure of the human ADP-ribosylation factor-1 complexed with GDP. Nature. 1994;372:704–708. - PubMed
-
- Austin C, Boehm M, Tooze SA. Site-specific cross-linking reveals a differential direct interaction of class 1, 2, and 3 ADP-ribosylation factors with adaptor protein complexes 1 and 3. Biochemistry. 2002;41:4669–4677. - PubMed
-
- Brodsky FM, Chen CY, Knuehl C, Towler MC, Wakeham DE. Biological basket weaving: Formation and function of clathrin- coated vesicles. Annu Rev Cell Dev Biol. 2001;17:517–568. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

