A conformational switch in PRP8 mediates metal ion coordination that promotes pre-mRNA exon ligation
- PMID: 23686287
- PMCID: PMC3703396
- DOI: 10.1038/nsmb.2556
A conformational switch in PRP8 mediates metal ion coordination that promotes pre-mRNA exon ligation
Abstract
Splicing of pre-mRNAs in eukaryotes is catalyzed by the spliceosome, a large RNA-protein metalloenzyme. The catalytic center of the spliceosome involves a structure comprising the U2 and U6 snRNAs and includes a metal bound by U6 snRNA. The precise architecture of the splicesome active site, however, and the question of whether it includes protein components, remains unresolved. A wealth of evidence places the protein PRP8 at the heart of the spliceosome through assembly and catalysis. Here we provide evidence that the RNase H domain of PRP8 undergoes a conformational switch between the two steps of splicing, rationalizing yeast prp8 alleles that promote either the first or second step. We also show that this switch unmasks a metal-binding site involved in the second step. Together, these data establish that PRP8 is a metalloprotein that promotes exon ligation within the spliceosome.
Figures
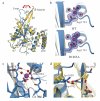
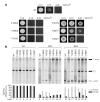
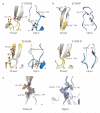

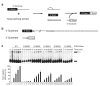

Comment in
-
Toggling in the spliceosome.Nat Struct Mol Biol. 2013 Jun;20(6):645-7. doi: 10.1038/nsmb.2603. Nat Struct Mol Biol. 2013. PMID: 23739164 No abstract available.
References
-
- Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell. 2003;12:5–14. - PubMed
-
- Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. - PubMed
-
- Madhani HD, Guthrie C. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell. 1992;71:803–817. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

